Although monitoring of select protein biomarkers has long been an integral part of standard oncology care (e.g., prostate specific antigen or carcinoembryonic antigen), methods with sufficient analytical sensitivity to enable detection of somatic tumor-associated variants from circulating tumor DNA (ctDNA) are now available. These methods have gained a critical place in solid tumor variant identification and monitoring and are now increasingly being applied to lymphomas. Historically, radiologic methods have been the standard of care for monitoring lymphoma responses to therapy (specifically positron emission tomography combined with computed tomography [PET/CT]), with histologic confirmation required in some circumstances. However, both histology and PET/CT are challenging in certain lymphoma types, especially classic Hodgkin lymphoma (cHL).
The initial application of ctDNA in lymphoma care was in the monitoring of clonotypic immunoglobulin gene rearrangements in diffuse large B-cell lymphoma (DLBCL) with a specificity of 100 percent (i.e., no false positives), versus only 56 percent for PET/CT.1 Subsequent studies introduced the use of somatic tumor variants for this purpose.2,3 Now, Drs. Valeria Spina and Alessio Bruscaggin and colleagues have demonstrated the utility of this method in cHL. Since prior studies were all performed on tumors with diffuse involvement by large numbers of lymphoma cells, the validity of this approach in a tumor type with rare neoplastic cells in a predominant inflammatory background is even more astounding. Drs. Spina and Burscaggin’s study makes several key points, as outlined in the following sections.
ctDNA identifies tumor-specific variants in cHL. Mutations identified in the ctDNA were compared to paired tumor gDNA, with 87.5 percent detection in ctDNA of variants identified in the tumor. None of these variants were detected in paired tumor background tissue without neoplastic cells, confirming their origin in the neoplastic cells. Additionally, all germline variants were excluded though comparison with paired normal gDNA from granulocytes. Additional variants found in ctDNA and not in the tumor (106 in ctDNA vs. 96 from tumor gDNA) tracked with disease status, confirming their assessment of neoplasia. Overall, somatic mutations were identified in 81.2 percent of cHL cases, with a mean of five mutations per case.
ctDNA tracks with tumor burden in cHL. The average variant allele fraction (VAF) was 5.5 percent, with most variants found at a VAF greater than 1 percent. Summing the ctDNA burden, there was correlation between the ctDNA load and Ann Arbor stage I versus stages II to IV (p = 0.021), limited (IA, IB, or IIA without bulky disease) versus advanced stage (III, IV, or I or II with bulky disease, or IIB; p = 0.001), and German Hodgkin Study Group Score favorable versus intermediate and unfavorable risk groups (p = 0.013).
Mutations can be found in key pathways with a predominance of STAT6 hotspot variants in cHL. STAT6 was the most commonly mutated gene (37.5% of cases) in this study as well as the concurrent study by Dr. Enrico Tiacci and colleagues (32% of cases).4 Two hotspot mutations in STAT6 (p.N417Y and p.D419N/H/A) among other variants are all located in the DNA binding domain and seem to support growth of cHL cells in a manner sensitive to JAK2 inhibitors.4 STAT6 mutations were associated with younger age, nodular sclerosis histologic subtype, lack of Epstein-Barr virus, and increased number of mutations. Dr. Tiacci and colleagues found that the JAK-STAT pathway was implicated in overall 87 percent of cHL cases.4 Other recurrent pathways included the NF-kB (46.2% of cases), PI3K/AKT (46.2%), epigenetic (35.0%), immune surveillance (27.5%), and NOTCH (20.0%) pathways. This pathway distribution is distinct from DLBCL but similar to that seen in primary mediastinal large BCL.
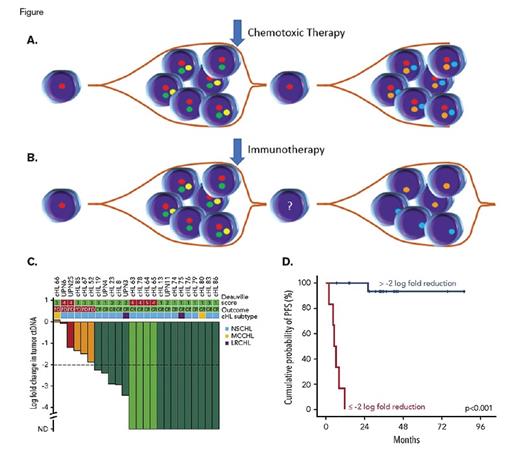
(A) Schematic of clonal evolution in cHL after chemotoxic therapy as determined by ctDNA analysis, with resurgence of the founder clone. (B) Schematic of clonal evolution in cHL after immunotherapy as determined by ctDNA analysis, with emergence of new clone(s). (C) Reduction in ctDNA burden correlates with likelihood of complete response. (Reprinted from Spina V, Bruscaggin A, Cuccaro A, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131:2413-2425.) (D) Probability of progression free survival (PFS) based upon a threshold of 2 log reduction in ctDNA burden. (Reprinted from Spina V, Bruscaggin A, Cuccaro A, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131:2413-2425.)
(A) Schematic of clonal evolution in cHL after chemotoxic therapy as determined by ctDNA analysis, with resurgence of the founder clone. (B) Schematic of clonal evolution in cHL after immunotherapy as determined by ctDNA analysis, with emergence of new clone(s). (C) Reduction in ctDNA burden correlates with likelihood of complete response. (Reprinted from Spina V, Bruscaggin A, Cuccaro A, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131:2413-2425.) (D) Probability of progression free survival (PFS) based upon a threshold of 2 log reduction in ctDNA burden. (Reprinted from Spina V, Bruscaggin A, Cuccaro A, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131:2413-2425.)
ctDNA reflects two different paths of clonal evolution in the setting of refractory disease in cHL. Interestingly, refractory cases that received chemotoxic therapy retained their founder clones (predominantly STAT6, TNFAIP3, ITPKB, and GNA13) with acquisition of new subclonal events, resulting in a trend toward an increased percentage of mutations in genes such as ARID1A, TET2, BCOR, SPEN, EP300, KMT2D, and XPO1 (Figure 1A). By contrast, those patients who received immunotherapy effectively cleared their founder clones with emergence of new clonal events (Figure 1B).
ctDNA predicts response to therapy in cHL. Similar to prior studies in DLBCL, the researchers suggest an optimized cutoff of a two-log reduction in ctDNA load in response to chemotoxic agents to predict complete response and cure (p < 0.001) (Figures 1C and 1D). By contrast, PET/CT in cHL is plagued by the fact that glucose uptake largely measures the inflammatory component of cHL and not the tumor burden. These results suggest that ctDNA may complement (or even ultimately replace) PET/CT monitoring.
In Brief
In summary, this study showcases the utility of ctDNA monitoring in cHL. This methodology highly correlates with the tumor genetics and burden, measures only the neoplastic cells and not the background inflammation, elucidates tumor biology and clonal evolution, and predicts outcome. In a disease where both morphologic and radiographic methods are plagued by the paucity of neoplastic cells and overwhelming inflammation, this targeted assessment of just the neoplastic cells through a minimally invasive method with a replenishing source of ctDNA may be the ideal method for monitoring these patients in the future.
References
Competing Interests
Dr. Kim indicated no relevant conflicts of interest.