CR, complete response; CR1, first CR; CR2, second CR; ELN, European Leukemia Network criteria; HSCT, hematopoietic stem-cell transplantation; NK, natural killer.
CR, complete response; CR1, first CR; CR2, second CR; ELN, European Leukemia Network criteria; HSCT, hematopoietic stem-cell transplantation; NK, natural killer.
The drive toward personalized medicine is predicated on the belief that the most effective treatment strategy can be reliably selected for an individual based on precise diagnostic and prognostic information. Currently though, for diseases such as acute leukemia, lymphoma, and myeloma, we typically practice “group medicine,” where we assign individuals to prognostic groups that dictate treatment choices that are uniform within the group. This approach has delivered improvements at the whole population level for patients with selected leukemia (think pediatric acute lymphoblastic leukemia), some lymphomas, and in myeloma. Moving to personalized medicine requires that we tackle the heterogeneity of disease within the “groups,” and currently, genomics is the favored tool for this task. Leading the charge is acute myeloid leukemia (AML), a genomically heterogeneous disease with restricted curative treatment options, but ever-increasing sophistication in diagnosis and well-established prognostic algorithms based on clinical features and genetic features.
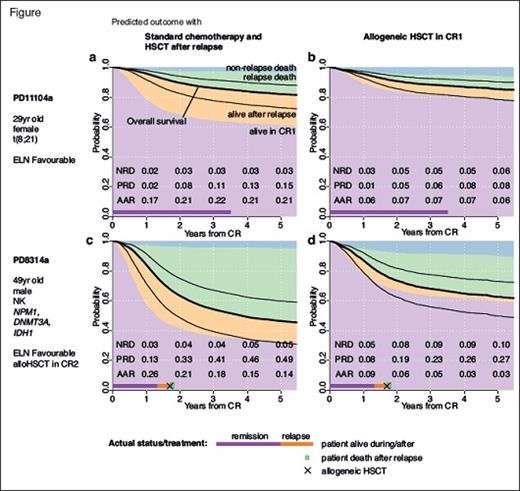
A major insight into how we can move from group medicine to personalized medicine came with the publication this year of “Precision Oncology for Acute Myeloid Leukemia Using a Knowledge Bank Approach” by Dr. Moritz Gerstung and senior authors Professors Peter Campbell and Harmut Doehner.1 In a seminal collaboration between the Wellcome Trust Sanger Institute in the U.K. and the German-Austrian AML Study Group, clinical data from three prospective trials of intensive therapy including 1,540 patients and matched genomic data on 111 genes for each patient were reanalysed to construct a multistage statistical model that accurately predicted likelihoods of remission, relapse, and mortality. After validation using data from The Cancer Genome Atlas, the authors tested the utility of the knowledge bank for generating personalized predictions tailored to the individual patient and how these predictions could be used as decision support for the choice between having an allogeneic stem cell transplant in first remission (CR1) or reserving this for after relapse.
The results are both exciting and sobering. First, this complex model encompassing 231 predictor variables outperformed current standard prognostic criteria such as those of the European Leukaemia Network (ELN) in predicting individual survival outcomes. It re-stratified approximately one-third of patients whose survival predictions deviated more than 20 percent from their ELN stratum. Thus, more information enabled substantially more accurate prediction of outcomes for individuals. Second, the knowledge bank could be used to predict the three-year probabilities of survival for patients who achieved CR1 whether they underwent an elective allograft in CR1 or not. Again, the model identified significant proportions of patients where the decision to allograft would be different based on standard criteria, including a subset of ELN favorable-risk patients who would likely improve their survival by more than 5 percent with an allograft, and a major subset of adverse risk patients where an allograft in CR1, as is typically recommended as standard, would not improve outcomes.
Based on this, the authors argued that by using the knowledge bank as a decision-support tool for individual patient treatment recommendations, allografts in CR1 could be reduced by 20 to 25 percent without worsening overall outcomes. Their economic modelling suggested that this would be cost saving, even after considering the costs of the genomic analyses.
The article also highlighted some daunting ongoing challenges should such an approach be implemented. Despite the size of this dataset, it is underpowered. The authors estimate that the survival predictions have a standard error of 6 percent, which is a level of uncertainty too high for use in many clinical circumstances. If data on 10,000 individuals could be included in the knowledge bank, an average prediction error for an individual patient survival estimate could be reduced to 2 percent. At that level of precision, it would be highly suitable for use in decision support. An international effort is ongoing to build the bank to this type of scale. To be appropriate for direct clinical use, the algorithm should also be prospectively validated, and genomic data should be generated using methodology accredited for clinical use. Although the knowledge-bank approach has the potential to change treatment decisions in a significant minority of patients, its routine application will not dramatically improve overall survival statistics for the populations of AML patients undergoing intensive treatment. The authors estimate its impact at that level would be an increase of approximately 1.3 percent, highlighting the ongoing unmet need for new therapies to drive big improvements in survival.
Dr. Gerstung and colleagues have shone a bright new light on the path we need to take to personalize decision making for our patients. More data are required, and knowledge banks must include patients who are representative of the wider population beyond clinical trial sets to enable meaningful extrapolation to real-world practice. This suggests the importance of building systems to incorporate data from patients undergoing routine clinical care into knowledge banks. Truly personalized decisions around major interventions such as transplantation will always require additional refinement to data-driven recommendations to enable incorporation of patient preference and the effect of confounders such as comorbidities, which may be dynamic in nature.
Knowledge banks also have the potential to help evaluate the impact of new therapies but will depend on quality clinical trials to generate the essential data, and Dr. Gerstung and colleagues have emphasized that large numbers of patients will be required to elucidate robust interactions between genetic aberrations and drug treatment effects. To enable personalized decisions when a variety of new or experimental therapies are possible, additional variables need to be analyzed. As Dr. Anthony Letai has reminded us,2 it’s not all about genomics, and those functional data on the effect of drugs on tumors may provide key clues for patients once their journey has moved beyond standardized initial therapy. With that in mind, an honorable mention goes to Dr. Jeffrey W. Tyner and colleagues for their efforts.3 They performed ex vivo sensitivity profiling of 122 primary patient samples from a variety of hematologic malignancies against a panel of 48 drug combinations and sought to incorporate this with genetic and phenotypic data to enable the rational development of new combinations of therapies. While at this point such efforts are designed to inform trial design and “group” medicine, they do provide additional illumination of how treatment choices may be made in the future, particularly for patients with advanced disease where trial-based evidence is lacking or not applicable.
These are two exemplars of the advances hematology made in 2017 in the quest to develop and deliver precision approaches that enable more personalized medical care. Systematic integration of clinical data (both trial and real world) with –omic and functional data seems to be the path to success. We should expect more light on this path in 2018 and the years that follow.
References
Competing Interests
Dr. Roberts indicated no relevant conflicts of interest.