The Question:
What genetic markers and risk stratification algorithms do you use to guide individualized treatment of juvenile myelomonocytic leukemia (JMML)?
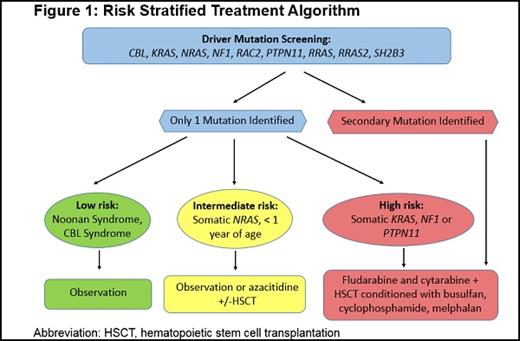
Figure 1. Risk Stratified Treatment Algorithm. Abbreviation: HSCT, hematopoietic stem cell transplantation.
Figure 1. Risk Stratified Treatment Algorithm. Abbreviation: HSCT, hematopoietic stem cell transplantation.
Our Response:
JMML is a rare and aggressive myelodysplastic/myeloproliferative neoplasm of infants and young children. It is characterized by leukoerythroblastosis on peripheral blood examination and fewer than 20 percent blasts in the bone marrow. Clinical features frequently include hepatosplenomegaly, fevers, rash, poor growth, and susceptibility to infections. Several congenital syndromes predispose to JMML, including neurofibromatosis type 1 (NF1) and CBL syndrome.1 Genetic mutations in NF1, NRAS, KRAS, RRAS, RRAS2, PTPN11, and CBL are all hypothesized to increase signaling through the Ras/MAPK pathway, which is a cardinal molecular feature that can be identified in approximately 95 percent of patients.2-4 Additionally, patients with Noonan syndrome and germline PTPN11 mutations can also develop a JMML-like myeloproliferative disease during childhood. Interestingly, patients with germline PTPN11 and CBL mutations who develop a MPN have high rates of spontaneous regression.5 Similarly, rare JMML patients with somatic NRAS and KRAS mutations experience spontaneous resolution of their disease.6 Unfortunately, identifying these patients at diagnosis has proven exceptionally difficult. In contrast to patients who experience self-remitting disease, overall outcomes in this disease after hematopoietic stem cell transplantation (HSCT) are quite poor, with event-free survival of less than 50 percent, primarily due to relapse.7 Updated diagnostic criteria for JMML, which are included in the revised 2016 World Health Organization classification, consist of a combination of clinical, laboratory and genetic/cytogenetic features (Table).8,9
How do you risk stratify JMML?
Recently, whole exome sequencing has identified additional genetic alterations contributing to relapse, including upregulation of JAK-STAT signaling, epigenetic modification through the PRC2 components such as DNMT3A, EZH2 and ASXL1, and altered RNA splicing.3,4 Mutations in a hotspot region of SETBP1 appear to be the single most common cause of relapse.10 Monosomy 7 is the most frequent cytogenetic abnormality, occurring in approximately 25 percent of patients with JMML. Several groups have recently demonstrated that an increased number of somatic alterations correlates with a poor prognosis in JMML.2-4
In patients who meet criteria for JMML, we stratify based on age, driver mutation, and the presence of secondary genetic alterations including monosomy 7. Given the high rate of spontaneous resolution in patients with CBL syndrome, these patients frequently can be observed without therapy. Intervention with chemotherapy is typically only indicated for CBL patients who become symptomatic (splenomegaly, failure to thrive, recurrent infections, etc.). Patients harboring NRAS mutations, particularly infants without secondary genetic alterations, can be closely monitored for signs of spontaneous resolution or treated with low-dose chemotherapy. Some case reports have also utilized azacytidine.11 All other patients should be considered for chemotherapy and then proceed to HSCT with the best available donor.
What conventional treatment approaches are employed in JMML?
One unresolved question in JMML is the role of pretransplant chemotherapy. In nearly every other pediatric leukemia, it is known that a lower disease burden at the time of transplantation is associated with improved outcomes. However, in JMML, many clinicians proceed directly to HSCT without decreasing disease burden with pretransplant chemotherapy. Another common treatment approach is to bridge to HSCT with oral 6-mercaptopurine or low-dose cytarabine.9 This approach reduces spleen size and white blood cell count but rarely reduces the allelic fraction of the underlying mutation (unpublished data). In contrast, we have observed that more intensive treatment with fludarabine and high-dose cytarabine is capable of eradicating somatic mutations by deep sequencing prior to HSCT. Whether this leads to improved long-term outcomes is still unknown, but our institutional bias is to offer this well-tolerated regimen to patients planning to undergo HSCT.
A recent clinical trial, Children’s Oncology Group (COG) ASCT1221 (NCT01824693), randomized patients to receive HSCT conditioning with busulfan and fludarabine, or busulfan, cyclophosphamide, and melphalan. It was closed prematurely after patients treated with busulfan and fludarabine were noted to have increased rates of disease progression. We therefore recommend using busulfan, cyclophosphamide, and melphalan for all patients undergoing HSCT until we can identify patients appropriate for a reduced-toxicity regimen.
JMML Diagnostic Criteria
| 1. Clinical and hematologic features (all 4 features mandatory) . |
|---|
|
| II. Genetic studies (1 finding sufficient) . |
|
| III. For patients without genetic features, besides the clinical and hematologic features listed under I, the following criteria must be fulfilled: . |
|
| 1. Clinical and hematologic features (all 4 features mandatory) . |
|---|
|
| II. Genetic studies (1 finding sufficient) . |
|
| III. For patients without genetic features, besides the clinical and hematologic features listed under I, the following criteria must be fulfilled: . |
|
Abbreviations: BM, bone marrow; CBL, Casitas B-lineage lymphoma; GM-CSF, granulocyte macrophage colony-stimulating factor; JMML, juvenile myelomonocytic leukemia; NF1, neurofibromatosis type 1; PB, peripheral blood; STAT5, signal transducer and activator of transcription 5.
*Germline mutations (including Noonan syndrome) need to be excluded.
†Occasional cases with heterozygous splice site mutations.
What is the role of novel targeted therapies in the current treatment algorithm?
There are two novel approaches currently in clinical trials in the United States and Europe. Nearly 100 percent of JMML patients harbor a Ras pathway alteration. Preclinical models using genetically engineered mice with Ras pathway mutations suggest that MEK inhibition could be an effective approach in this disease.12 The Children’s Oncology Group will therefore be sponsoring a clinical trial (ADVL1521) to test the efficacy of the oral MEK inhibitor, trametinib, in patients with relapsed and refractory JMML. As all monotherapy regimens are likely to be limited by the development of resistance, one of the secondary objectives of this trial is to identify mechanisms of resistance to rationally plan future combinatorial treatments.
There are several case reports of complete remissions achieved with the hypomethylating agent azacytidine in JMML.11 In Europe, there are ongoing clinical trials testing the safety and efficacy of azacytidine in patients with newly diagnosed and relapsed/refractory JMML.
What are future directions for treatment and what questions remain?
Several additional agents that are undergoing preclinical testing include Ras mimetics, SHP-2 antagonists, anti-GM-CSF antibodies, and CAR-T cells targeting JMML-specific antigens.
The optimal pretransplant therapy is likely to be a combination of cytotoxic chemotherapy, MEK inhibition, hypomethylation agents, and other targeted agents. It is important to note that there has never been a randomized trial of pretransplant regimens in JMML. Due to the rarity of this disease, an international collaborative trial is the only feasible approach to carrying out such a trial. Recently, response criteria have been proposed for evaluating outcomes to non-transplant therapies and HSCT in the context of clinical trials, which should help with comparison of different treatment modalities.13 Lastly, as disease courses vary dramatically from spontaneous resolution to rapid progression after HSCT, a robust risk-stratification algorithm based on multiple biomarkers will be critical to predict these divergent outcomes and inform treatment decisions for patients with JMML.
References
Competing Interests
Dr. Elliot Stieglitz and Dr. Ashley Koegel indicated no relevant conflicts of interest.