In the past decade, studies using advanced high-throughput sequencing technologies have provided insights into the genetic basis of myelodysplastic syndromes (MDS). This work has confirmed that 1) the bone marrow cells in patients with MDS (regardless of blast count) are clonally derived; 2) founder and multiple subclones exist simultaneously (i.e., clonal heterogeneity);1,2 and 3) mutations in several genes predict poor overall survival in patients independent of established clinical risk factors.3,4 Several of these studies have attempted to define the clonal architecture of MDS and the genetic changes that underlie its progression to secondary acute myeloid leukemia (sAML). This work has largely been restricted to studying the allelic burden of a limited number of driver mutations at a single time-point or performing comprehensive mutation profiling of serial samples from a small number of patients,1,2 potentially obscuring the clinical utility of this type of testing in diagnosis and dynamic prognostication and therapy selection.
To further clarify the relationship between molecular changes and disease progression and clinical outcomes in MDS, Dr. Hideki Makishima and colleagues analyzed somatic mutations in a very large cohort of MDS patients, including many serially collected patients. Clonal architecture and dynamics were investigated by whole exome sequencing (WES) and/or targeted sequencing of 699 patients with MDS and sAML; 122 patients were analyzed longitudinally. These and published data sets (n=2,250) were interrogated to determine the differential roles of driver mutations in disease progression. They found that higher-risk MDS was characterized by a higher number of mutations with increasing diversity and a larger clone size. Additionally, the number of cases with heterogeneity was significantly higher in sAML compared with lower-risk MDS.
In WES analysis of serial samples, evolution of a new dominant clone that swept out other subclones was common during disease progression and was frequently accompanied by newly emerging subclones. In most cases, leukemic transformation was marked by acquisition of new mutations without clone sweeping.
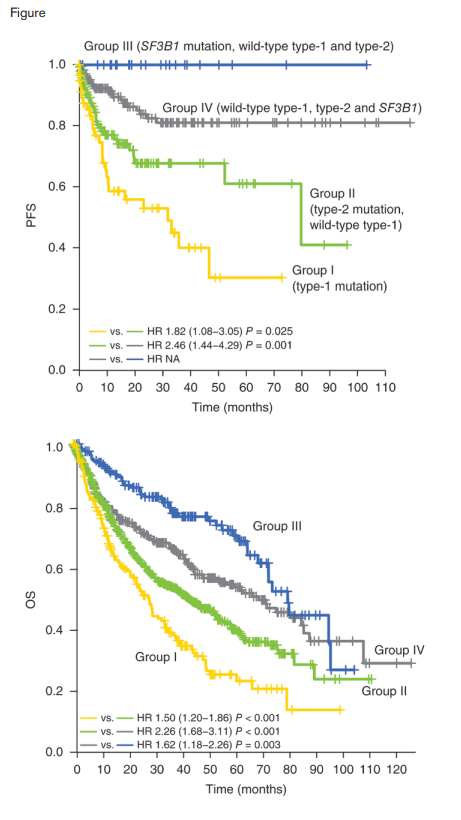
Progression-free and Overall Survival Curves. Kaplan-Meier curves for progression free survival (n = 429) A) and overall survival (n = 1,347) B) of patients with type 1 mutations (group I), with type 2 but not type 1 mutations (group II), with SF3B1 but no type 1 or type 2 mutations (group III), and other patients with no type 1, type 2 or SF3B1 mutations (group IV). Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics doi:10.1038/ng.3742, copyright 2016.
Progression-free and Overall Survival Curves. Kaplan-Meier curves for progression free survival (n = 429) A) and overall survival (n = 1,347) B) of patients with type 1 mutations (group I), with type 2 but not type 1 mutations (group II), with SF3B1 but no type 1 or type 2 mutations (group III), and other patients with no type 1, type 2 or SF3B1 mutations (group IV). Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics doi:10.1038/ng.3742, copyright 2016.
To clarify the relationship between driver mutations and disease progression, the authors compared the frequencies of major driver mutations in low- and high-risk MDS, as well as sAML. Mutations in genes designated as type 1 (FLT3, PTPN11, WT1, IDH1, NPM1, IDH2, and NRAS) were significantly enriched in sAML compared with high-risk MDS. When they compared high- and low-risk MDS, mutations in GATA2, KRAS, TP53, RUNX1, STAG2, ASXL1, ZRSR2, and TET2 (designated type 2 mutations) were significantly enriched in high-risk MDS. SF3B1 mutations were strongly enriched in low-risk MDS, compared to high-risk MDS. In longitudinal samples, type 1 mutations were more likely to be newly acquired or to increase in clone size. In contrast, more type 2 and other mutations decreased in their clone size or were even lost in the second sampling. As shown in the Figure, patients with type 1 mutations (group I) had a significantly shorter time to progression to sAML compared to patients with type 2 mutations who lacked type 1 mutations (Group II) and those lacking type 1 and type 2 mutations (group IV). Patients with SF3B1 mutations and lacking type 1 and type 2 mutations (group III) demonstrated a very low rate of leukemic progression. Overall survival of group I patients was significantly shorter than that of group II patients, who had a significantly shorter survival than group IV patients. In a multivariate analysis, the group I category remained an independent negative predictor of overall survival, together with other factors, including complex karyotype, IPSS score, −7/del(7q), age, and del(20q). SF3B1 mutations were mutually exclusive of other splicing factor mutations, and type 1 and type 2 mutations, potentially reflecting unique biology among this clinically distinct MDS subtype.5 They observed a global trend of co-occurrence of type 1 and type 2 mutations, with TP53, TET2, and NPM1 acting as exceptions and present largely mutually exclusive of other type 1 and type 2 mutations. Interestingly, type 1 mutations had significantly lower variant allele fractions compared to type 2 mutations in the patients with both categories of mutations, suggesting that type 1 mutations were acquired during disease progression after previous type 2 hits.
In Brief
In demonstrating that MDS disease progression is accompanied (and potentially mediated) by distinct molecular events, this work suggests that screening for these specific mutations might be useful to predict clinical outcome or allow therapeutic intervention at critical time points. Proof of this clinical utility requires further study, including elucidation of the underlying pathophysiology represented by these genetic findings.
References
Competing Interests
Dr. Keel indicated no relevant conflicts of interest.