Acute myeloid leukemia (AML) is generally divided into prognostic subgroups based primarily on cytogenetic alterations, with further refinement based on a small group of molecular markers, and the presence of morphologically identifiable residual disease after treatment.1 Half of AML cases in young adult patients fall into the standard-risk subgroup based on cytogenetic findings, with most cases showing a normal karyotype. This group is prognostically diverse, with some patients benefiting from bone marrow transplantation in first remission, and others following a low-risk disease course. New parameters to improve prognostic grouping in these patients would provide a valuable addition to treatment planning. Molecular alterations can help to further subdivide this group based on the presence of insertion mutations in NPM1 (nucleophosmin), and FLT3 (Fms-like tyrosine kinase 3) internal tandem duplications (ITD). Although DNMT3A (DNA methyltransferase 3A) mutations have been associated with adverse outcomes in normal karyotype AML, various studies indicate that prognosis may depend on patient age, type of DNMT3A mutation (R882 vs. non-R882), and the concurrent mutational status of NPM1 and FLT3.2-5
NPM1 mutations are leukemia-specific markers not seen in normal cells and are therefore of interest as targets for the detection of minimal residual disease (MRD) following therapy. MRD assessment is commonly used in acute lymphoblastic leukemia; however, reliable molecular markers in AML have been more difficult to identify due to the heterogeneity of the disease.6 Previous studies have demonstrated the utility of cytogenetic abnormalities or flow cytometric findings as markers for MRD.7,8 The utility of NPM1 mutation for MRD monitoring in the context of expanded molecular profiling by next-generation sequencing has not been demonstrated. Additionally, NPM1 mutations may not be stable during the course of disease, and a mutation identified at diagnosis may not be present at relapse.
To examine NPM1 mutation(s) as a tool for MRD monitoring, Dr. Adam Ivey and colleagues from the United Kingdom National Cancer Research Institute (NCRI) AML Working Group studied the presence of NPM1 mutation(s) at diagnosis, after each cycle of treatment, and quarterly for 24 months following consolidation therapy in 346 patients enrolled on the NCRI AML17 trial. Clinicians were not informed about the results of residual disease testing. To identify mutated transcripts in bone marrow and peripheral blood samples, they used a highly sensitive reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR) assay using mutation-specific primers for the three most common NPM1 mutations. In patients with rare mutations, patient-specific primers were designed. Mutated transcript levels were compared to the expression of the ABL1 reference gene. Positivity was defined as amplification in at least two of three replicates at a cycle-threshold value of 40 or less. Molecular remission was defined as an absence of detectable NPM1-mutated transcripts by RT-qPCR in a bone marrow sample at a sensitivity of one in 10,000. Molecular relapse was defined as the detection of increasing levels of NPM1-mutated transcript in two successive samples. Additionally, samples were sequenced on the Illumina HiSeq 2000 using a custom, targeted, 51-gene panel based on published data for NPM1-mutated AML and by exome sequencing of 22 patients in the study cohort.
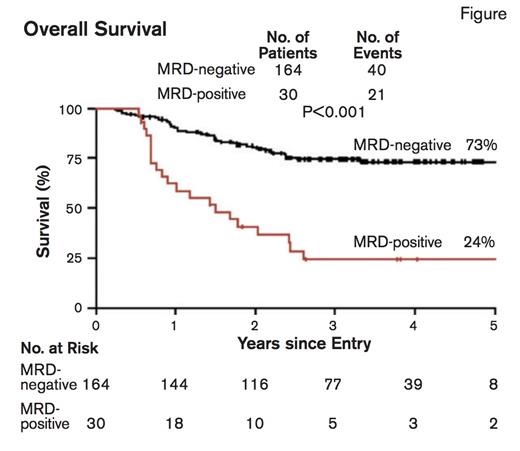
Overall Survival. From N Engl J Med, Ivey A et al, Volume 374, Page 422-433. Copyright © 2016 Massachusetts Medical Society. Reprinted with permission from Massachusetts Medical Society.
Overall Survival. From N Engl J Med, Ivey A et al, Volume 374, Page 422-433. Copyright © 2016 Massachusetts Medical Society. Reprinted with permission from Massachusetts Medical Society.
The presence of MRD based on the amount of mutated NPM1 transcripts was highly informative. In patients with a morphologic remission, a rising NPM1 transcript level reliably predicted hematologic relapse. The persistence of mutated transcripts in peripheral blood following the second chemotherapy cycle was most informative and was associated with a significantly higher risk of relapse at three years than was the absence of such transcripts (82% vs. 30%; p<0.001). Additionally, the five-year overall survival rate for MRD-negative versus MRD-positive patients was 73% versus 24%; p<0.001 (Figure). No mutation profile identified in the 51-gene panel sequencing results demonstrated a significant difference in survival or risk of relapse compared with the presence of MRD. Patients with MRD were more likely to have the FLT3-ITD mutation or a high Medical Research Council (MRC) risk score. The presence of MRD emerged as the sole prognostic factor for relapse in a multivariate analysis. Among patients considered high-risk due to FLT3-ITD, DNMT3A mutation, or both, the absence of mutated NPM1 transcripts identified patients with a relatively favorable outcome with a survival rate of 76 percent. Furthermore, NPM1 mutation was in fact a stable marker of disease, and was detectable in 69 (99%) of 70 patients at the time of relapse. Notably, DNMT3A and IDH mutations, molecular abnormalities associated with preleukemic clones, persisted at high levels in patients during long-term remission.
To further assess the use of quantitative NPM1 mutation analysis in MRD, a validation cohort of 91 patients with mutations was studied prospectively. Clinicians were informed of the results of MRD testing in this group. This analysis confirmed the association of MRD with a significantly worse outcome at two years due to increased incidence of relapse (70% vs. 31%; p=0.001) and lower rate of overall survival (40% vs. 87%; p=0.001). Stem cell transplantation was performed in 21 of 46 patients with MRD and in 61 of 239 patients without MRD. The absence of MRD remained prognostic in patients receiving stem cell transplantation.
In Brief
This study demonstrates the value of using highly sensitive RT-qPCR–based methods to identify MRD in patients with NPM1-mutated AML. Patients with intermediate-risk AML based on normal karyotype, presence of NPM1 mutation, and absence of FLT3-ITD and DNMT3A mutation are generally considered to have a better prognosis. Screening for MRD can help identify patients in this group who have a higher risk of relapse and poor outcomes.9 Further studies are necessary to explore the benefit of transplantation or more intensive chemotherapy in these patients. Patients with intermediate-risk AML and presence of FLT3-ITD and/or DNMT3A mutation without MRD identified by the absence of mutated NPM1 transcript after the second chemotherapy cycle may avoid stem cell transplantation in first remission. These results show that in addition to its role in risk stratification, NPM1 mutation can be used for MRD monitoring.
References
Competing Interests
Dr. Chabot-Richards and Dr. George indicated no relevant conflicts of interest.