Dr. Ray Schofield first hypothesized the concept of a hematopoietic stem cell (HSC) niche in 1978.1 He proposed that:
The stem cell is seen in association with other cells which determine its behavior… its maturation is prevented, and as a result, its continued proliferation as a stem cell is assured. Its progeny, unless they can occupy a similar stem cell “niche,” are first generation colony-forming cells [progenitors].
This hypothesis implies that stem cells are localized within specific locations in the bone marrow and that these locations govern the function of stem cells, and in particular, their proliferation or quiescence. Work ever since, and even before Dr. Schofield’s hypothesis, has sought to determine the location of HSCs in vivo and to characterize their surrounding supportive “niche” cells.
Several technological advances throughout the last decade have now allowed for the visualization of presumed HSCs in tissue and have created numerous different models of the location and characterization of the HSC niche. Many studies on the hematopoietic niche in the last decade have relied on antibody staining of phenotypic markers known to enrich for HSCs, most notably the SLAM family of markers in mice (CD150+, CD48-, CD244-).2 This earlier report2 of HSC localization using SLAM markers was performed on bone sections from three femurs. The authors were able to identify 35 total cells, allowing for some speculation about HSC localization, but a relatively modest number of total events precluded definitive conclusions.
Later, advances in confocal imaging coupled with phenotypic labeling of HSCs allowed for live imaging of HSCs in a transplantation scenario.3 These experiments demonstrated that repopulating HSCs specifically trafficked to, and engrafted near the endosteum in irradiated mice, suggesting that this endosteal surface was a potential HSC niche location. However, this early study was also performed with a modest number of events and was limited by the penetration depth of the imaging. The results also reflected HSCs that were transplanted, rather than endogenous HSCs at steady state.
A few years ago, an innovative method using whole-mount confocal imaging of the mouse sternum, coupled with 3D-rendering and computational analysis, demonstrated that quiescent HSCs were localized near bone marrow arterioles at a much higher rate than at random locations.4 In contrast to the previously described studies, the authors successfully imaged hundreds of HSCs in bones per study, and remarkably the same numbers of HSCs were detected in the whole-mount imaging as by flow cytometry — a significant improvement over prior studies.
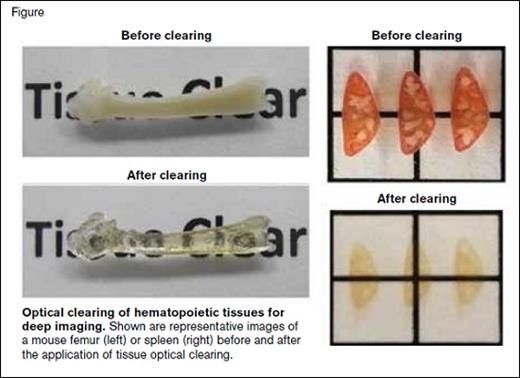
Optical Clearing of Hematopoietic Tissues for Deep Imaging. Shown are representative images of a mouse femur (left) or spleen (right) before and after the application of tissue optical clearing.
Optical Clearing of Hematopoietic Tissues for Deep Imaging. Shown are representative images of a mouse femur (left) or spleen (right) before and after the application of tissue optical clearing.
A new study by Dr. Melih Acar and colleagues has now added to these technological advances in imaging HSCs in tissue — application of tissue optical clearing and a new endogenous reporter mouse. Expanding upon whole-mount imaging techniques, the authors used a tissue-clearing technique previously employed in brain tissues or mouse embryos, to create “clear” bones (Figure), allowing for deeper penetration of confocal imaging.
Using gene expression profiling from their earlier studies that identified the SLAM markers,2 the authors also found that α-catulin was highly expressed in HSCs compared with unfractionated bone marrow cells. A green fluorescent protein (GFP) was then inserted into the first exon of α-catulin, creating a reporter mouse in which approximately 50 percent of the SLAM cells were GFP+, and there was no detection of GFP in more mature hematopoietic cells. Coupling GFP expression with antibody staining for c-kit led to similar enrichment for long-term repopulating HSCs as SLAM markers. Using this two-marker system and optically cleared sections, the authors used similar spatial analysis to the prior report4 and demonstrated that these endogenously GFP-labeled c-kit+ cells were contained more commonly within the central marrow rather than in bone surfaces. Approximately 85 percent of the cells were within 10 μm of a sinusoidal vessel. However, if a similar analysis was performed by comparing the distance of randomly placed spots in the bone marrow cavity to sinusoids, there was no statistical difference between the random spots and the HSCs, due to the abundance of sinusoids in the bone marrow space. Surprisingly, there was no localization difference observed between cycling versus noncycling HSCs. The authors also coupled their imaging analysis with CXCL12 reporter mice and Leptin-receptor reporter mice and demonstrated that the vast majority of HSCs are within 5 μm of these reporter cells within the marrow niche. While this localization of HSCs was significantly different than randomly placed spots, 85 to 90 percent of random spots also were within 5 μm of these reporter cells, demonstrating how ubiquitous these cells and “niche” locations are within the marrow space.
Recently, a number of studies have described numerous cell types that comprise the HSC niche, largely driven by the availability of a specific Cre-recombinase or related reporter mouse strain. Consequently, a series of publications have described the “cell identity” of the HSC niche based on the mouse model including Lepr-Cre, Prx1-Cre, Nestin-Cre, NG2-Cre, CXCL12-GFP, Mx1-Cre, Osx-Cre, and others. While the field has advanced as a result of these new mouse models, many of these Cre systems were not conditionally activated, meaning that the phenotype observed may not faithfully represent what happens in the adult stem cell niche. Secondly, none of these Cre or reporter systems are restricted to an exclusive cell type, and there are many known and unknown overlaps. Finally, different variants of the same reporter system often result in different phenotypes, adding to uncertainty in niche cell identity.
In Brief
The next decade of HSC niche research is likely to involve the use of still-emerging technologies, and our understanding of the niche (specifically what has occurred this past decade) will become simultaneously clearer and more convoluted. It will be important to apply these technologies both in steady-state situations as well as in situations of stress and disease, particularly HSC transplantation. Notably, optical clearing and deep tissue imaging also has been used by the authors to begin to explore HSC localization in the spleen (Figure) during extramedullary hematopoiesis.5 As more groups adopt these new techniques and apply them to their own model systems it is likely that the HSC niche will continue to be parsed into smaller and smaller subsets of locales and cells, perhaps with differing regulatory properties.
References
Competing Interests
Dr. Hoggatt indicated no relevant conflicts of interest.