Juvenile myelomonocytic leukemia (JMML) is a rare and unique form of childhood leukemia with both myelodysplastic and myeloproliferative features and clinical and biological similarities to chronic myelomonocytic leukemia (CMML) and chronic myelogenous leukemia (CML).1 The hallmark of this disease is hyperactive Ras signaling. While JMML often arises within the context of an inherited syndrome, de novo cases occur as well. Hematopoietic stem cell transplantation is presently the sole curative treatment option, but this has a success rate of only approximately 50 percent overall. There is great variability in the clinical course of JMML, and one of the most significant challenges in managing this disease is distinguishing children who will have favorable versus unfavorable outcomes.
To address these challenges, Dr. Elliot Stieglitz and colleagues performed a comprehensive genomic characterization of primarily nonsyndromic JMML, with the goal of identifying new mutations to refine outcome prediction and to aid in the development of new therapies. The authors first performed whole exome sequencing in 29 cases of JMML, comparing paired germline and diagnostic tumor tissue. Tumor samples from progression or relapse were analyzed in a subset of patients as well. The authors applied an innovative informatics algorithm to accurately distinguish somatic and germline mutations, which was of critical importance because approximately 25 percent of patients are known to have inherited syndromes that predispose to JMML.
Key findings from this analysis were the discovery of 10 new mutations in known oncogenes and tumor suppressor genes to add to the list of mutations in five canonical Ras pathway genes (NF1, KRAS, NRAS, PTPN11 and CBL), which have been previously implicated in the pathogenesis of JMML. These newly discovered genes are involved in diverse processes, including signal transduction, splicing, transcription, and epigenetic regulation, and they help introduce the possibility of treating JMML with existing targeted agents, such as Janus kinase (JAK) inhibitors or epigenetic agents. All of the identified pathogenic mutations were validated in an independent cohort of 71 patient samples using targeted deep sequencing.
Additional key findings in this report included the discovery of mutations in SH2B3 in a subset of JMML patients. Mutations in this tumor suppressor gene lead to JAK-STAT pathway activation. While mutations in SH2B3 have been observed in adult myeloproliferative neoplasms and lymphoid malignancies, this was the first report in JMML. The genomic complexity of JMML was further highlighted by the observation that coexisting mutations in Ras pathway and other genes were identified in 11 percent of patients, so not all Ras pathway alterations are mutually exclusive. Furthermore, mutations in epigenetic modifier genes were observed in 14 percent of patients and led to global hypermethylation in those studied. Finally, the authors illustrated several examples of the acquisition of secondary genetic events at the time of disease progression or relapse.
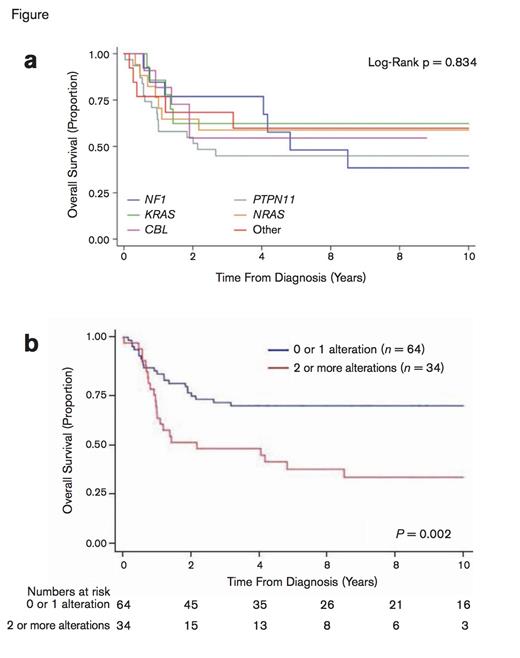
Patient Outcomes Stratified by Canonical Mutation and Number Of Somatic Alterations. (a) Overall survival based on canonical mutation. Individual canonical driver mutations were not associated with outcome (logrank p = 0.834). (b) Overall survival rates according to the number of somatic alterations at diagnosis. Adapted by permission from Macmillan Publishers Ltd: Nat Genet Volume 47 Pages 1326-1333, copyright 2015.
Patient Outcomes Stratified by Canonical Mutation and Number Of Somatic Alterations. (a) Overall survival based on canonical mutation. Individual canonical driver mutations were not associated with outcome (logrank p = 0.834). (b) Overall survival rates according to the number of somatic alterations at diagnosis. Adapted by permission from Macmillan Publishers Ltd: Nat Genet Volume 47 Pages 1326-1333, copyright 2015.
The authors next analyzed the prognostic contribution of genetic mutational profile. Notably, they observed that the number of somatic alterations at diagnosis rather than the specific mutation, determined prognosis (Figure). Overall survival rates at 10 years were significantly better (65.1% ± 6.0% vs. 29.0% ± 8.3%; P = .002) in the 65 percent of patients with zero or one somatic alterations at diagnosis, compared with the 35 percent of patients who had two or more mutations. Moreover, in a multivariate analysis, the number of somatic alterations at diagnosis was the most significant predictor of outcome, exceeding the prognostic impact of traditional clinical risk factors.
In Brief
There is great heterogeneity in the clinical course of JMML, with some children experiencing spontaneous disease regression while others exhibiting an aggressive and rapidly progressive course. Some of the greatest challenges in the management of children with JMML include this unpredictability in the clinical course as well as the limited number of effective treatment options. This study by Dr. Stieglitz and colleagues sheds important new light on the pathogenesis and genetic complexity of JMML and offers opportunities for both refinements in outcome prediction as well as the potential for development of novel combinational therapies, targeting both driver and acquired secondary mutations.
References
Competing Interests
Dr. Raetz indicated no relevant conflicts of interest.