The idea of a dystopic future in which mankind genetically engineers offspring for desirable traits or in order to make “super humans” has long been a focal point of many works of science fiction. At the beginning of this year, rumblings in the scientific community and media began to suggest that science fiction was quickly becoming reality. In January, a leading group of genetic scientists met in Napa, California, to discuss the implications of genomic engineering and published a joint opinion on the subject in Science in April.1 In that opinion, the authors recommended that steps be taken to
“…strongly discourage, even in those countries with lax jurisdictions where it might be permitted, any attempts at germline genome modification for clinical applications in humans, while societal, environmental, and ethical implications of such activity are discussed among scientific and governmental organizations.”
A similar piece was published in Nature2 a few days before from leading scientists at Sangamo and the Alliance for Regenerative Medicine titled “Don’t Edit the Human Germ Line.” Both groups cited that the technology was not yet adequately understood (particularly the long-term consequences) to warrant editing of human reproductive cells, and that public misperceptions about such efforts could undermine promising work on gene editing of somatic cells that has the potential to treat those with existing diseases. These efforts in gene therapy of adult cells, in relation to blood disorders, were highlighted in the January/February 2015 “Year’s Best” edition of The Hematologist.3
The prominent opinion pieces in Science and Nature were published in likely anticipation of a report in mid-April in the journal Protein Cell from Dr. Puping Liang and colleagues at Sun Yat-sen University in China. This was the first published report of genetic engineering of human embryos, and it garnered an intense amount of media coverage. One CBS news report on the manuscript referred to the study as the “Designer Baby Controversy,”4 a similar sentiment in many other media outlets.
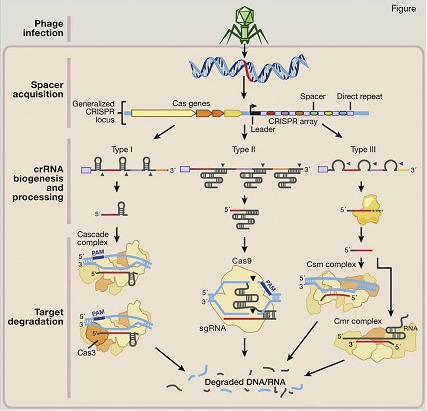
Natural Mechanisms of Microbial CRISPR Systems in Adaptive Immunity. Following invasion of the cell by bacteriophages or plasmids (step 1: phage infection), certain CRISPR-associated (Cas) enzymes acquire spacers from the exogenous protospacer sequences and install them into the CRISPR locus within the prokaryotic genome (step 2: spacer acquisition). These spacers are segregated between direct repeats that allow the CRISPR system to mediate self and nonself recognition. The CRISPR array is a noncoding RNA transcript that is enzymatically maturated through distinct pathways that are unique to each type of CRISPR system (step 3: crRNA biogenesis and processing). In types I and III CRISPR, the pre-crRNA transcript is cleaved within the repeats by CRISPR-associated ribonucleases, releasing multiple small crRNAs. In type II CRISPR, which was used by the authors of the zygote editing manuscript, an associated trans-activating CRISPR RNA (tracrRNA) hybridizes with the direct repeats, forming an RNA duplex that is cleaved and processed by endogenous RNase III and other unknown nucleases. Maturated crRNAs from type I and III CRISPR systems are then loaded onto effector protein complexes for target recognition and degradation. In type II systems, crRNA-tracrRNA hybrids complex with Cas9 to mediate interference.Reprinted from Cell, Vol. 157, Hsu et al, "Development and Applications of CRISPR-Cas9 for Genome Engineering," Pages 1262-1278, Copyright 2014, with permission from Elsevier.
Natural Mechanisms of Microbial CRISPR Systems in Adaptive Immunity. Following invasion of the cell by bacteriophages or plasmids (step 1: phage infection), certain CRISPR-associated (Cas) enzymes acquire spacers from the exogenous protospacer sequences and install them into the CRISPR locus within the prokaryotic genome (step 2: spacer acquisition). These spacers are segregated between direct repeats that allow the CRISPR system to mediate self and nonself recognition. The CRISPR array is a noncoding RNA transcript that is enzymatically maturated through distinct pathways that are unique to each type of CRISPR system (step 3: crRNA biogenesis and processing). In types I and III CRISPR, the pre-crRNA transcript is cleaved within the repeats by CRISPR-associated ribonucleases, releasing multiple small crRNAs. In type II CRISPR, which was used by the authors of the zygote editing manuscript, an associated trans-activating CRISPR RNA (tracrRNA) hybridizes with the direct repeats, forming an RNA duplex that is cleaved and processed by endogenous RNase III and other unknown nucleases. Maturated crRNAs from type I and III CRISPR systems are then loaded onto effector protein complexes for target recognition and degradation. In type II systems, crRNA-tracrRNA hybrids complex with Cas9 to mediate interference.Reprinted from Cell, Vol. 157, Hsu et al, "Development and Applications of CRISPR-Cas9 for Genome Engineering," Pages 1262-1278, Copyright 2014, with permission from Elsevier.
The researchers took advantage of the relatively new gene-editing tool, CRISPR/Cas9.5,6 CRISPRs (clustered regularly interspaced palindromic repeats) are short sequences in the genomes of several bacteria and archaea, and act as a form of adaptive immunity against viruses and plasmids. When exposed to a bacteriophage, short phage-related spacer sequences are incorporated into the CRISPR region of the bacteria genome. Then, when the bacteria are exposed to the phage again, this spacer sequence interacts with CRISPR-associated (Cas) nucleases and, similar to RNAi, binds to the specific phage sequence and cuts the DNA. In a series of recent studies, it was determined that the “guide RNA” could be programmed to cut any portion of the genome desired,7 including in mammalian cells.8-10 Unlike zinc finger nucleases (ZFNs) or transcription activator-like effector nucleases (TALENs), where the protein specifically recognizes DNA and hence requires complicated protein engineering, the single Cas9 endonuclease is the only protein needed, and genome specificity is generated through the guide RNA — a relatively simple synthesis procedure.5,11 This has opened the door of genome editing to a much broader group of scientific teams than the previous technologies.
Using CRISPR/Cas9 editing, Dr. Liang and colleagues set out to determine whether they could edit a portion of the human β-globin gene, which is mutated in β-thalassemia, in preimplantation embryos. The team used tripronuclear zygotes that form as a result of two sperm nuclei in one oocyte — an occurrence that happens roughly five percent of the time during in vitro fertilization (IVF) programs. The triploid zygotes are not able to develop normally and do not result in births, and the team stated that these embryos were specifically chosen to avoid ethical concerns. The team then co-injected 86 embryos with an optimized guide RNA to target the β-globin gene (found in prescreens in a cell line), the mRNA for Cas9 nuclease, green fluorescent protein (GFP) mRNA, and a ssDNA oligo, which was a template for the corrected β-globin gene. Of the 86 zygotes, 71 were living 48 hours after injection. Of those, 59 expressed GFP, and of those, 54 were able to be amplified by polymerase chain reaction for further evaluation. Within the 54 that were genetically tested, about half (28) of them were cleaved by Cas9, and of those, only four were successfully edited with the repair template. The authors also noted several off-target mutations as a result of the editing procedure; it was incomplete, as the resulting embryos were mosaic, containing multiple different versions of the same gene. The authors conclude in their discussion:
“Because the edited embryos are genetically mosaic, it would be impossible to predict gene editing outcomes through pre-implantation genetic diagnosis (PGD). Our study underscores the challenges facing clinical applications of CRISPR/Cas9.”
In Brief
Despite the media reports to the contrary, it is fairly clear from this report that “designer babies” are not being created in China. This manuscript has, however, ignited discussion regarding the ethical paths forward for genome editing. Shortly after publication, NIH director Francis Collins reiterated the government and agency policies against similar research in human embryos.12 The authors themselves may have outlined the larger question of embryo gene editing when they mentioned preimplantation genetic testing, which is already occurring clinically. Dr. Rudolf Jaenisch, president of the International Society for Stem Cell Research (ISSCR), was quoted in the New York Times, stating that, “with gene editing, the cutting and pasting has to start immediately… given that (in most cases of genetic disease) half the embryos that were edited would be normal — their DNA would have been forever altered for no reason.”13 Attention on gene editing should be focused at this time on therapies in somatic cells to treat children and adults with genetic diseases. Researchers exploring somatic cell gene editing for blood disorders are pioneering this field, and the hematologic scientific community should help inform the public about the differences between these ongoing efforts and those related to germline modification.
References
Competing Interests
Dr. Hoggatt indicated no relevant conflicts of interest.