It is indeed tragic that the consequence of the initial, potentially curative therapy of cancer can be a second, rapidly progressive fatal condition, therapy-related myelodysplastic syndrome (t-MDS), and/or acute myeloid leukemia (t-AML). t-MDS and t-AML are difficult to manage – from a psychological standpoint, as patients are now devastated by an aggressive additional malignancy after what may have been a long course of prior surgery, radiation, and/or chemotherapy; and from a therapy standpoint, as there are typically poor outcomes due to chemotherapy drug resistance. The highest rates of t-MDS/AML are observed after treatment of breast cancer or lymphoma. It has been assumed that because of chemotherapy- and radiation-induced DNA damage, t-MDS/AML patients would exhibit many more mutations compared to de novo AML, and perhaps more novel mutations as well. The most common mutated gene in t-MDS/AML is the TP53 (tumor protein p53) tumor suppressor gene.
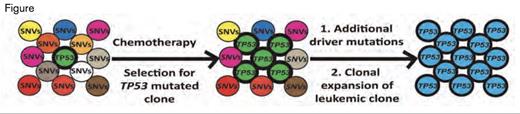
Development of t-AML Due to Clonal Expansion of the TP53-Mutated Clone. Chemotherapy imposes selection for the TP53 mutated clone leading to a higher representation (middle panel). Additional driver mutations lead to expansion of leukemic clones (right panel). SNVs=single nucleotide variants.Reprinted by permission from Macmillan Publishers Ltd: Nature. 2014 Dec 8. doi: 10.1038/nature13968, copyright 2014.
Development of t-AML Due to Clonal Expansion of the TP53-Mutated Clone. Chemotherapy imposes selection for the TP53 mutated clone leading to a higher representation (middle panel). Additional driver mutations lead to expansion of leukemic clones (right panel). SNVs=single nucleotide variants.Reprinted by permission from Macmillan Publishers Ltd: Nature. 2014 Dec 8. doi: 10.1038/nature13968, copyright 2014.
Dr. Terrence Wong, Dr. Giridharan Ramsingh, and Dr. Andrew Young and colleagues at Washington University in St. Louis have debunked prior theory on the origin of therapy-related hematopoietic disorders after performing whole genome sequencing of 22 cases of t-AML. They observed a similar rate of all types of mutations (single-nucleotide variants, small insertions or deletions [indels], and transversions [substitution of purine for pyrimidine or vice versa]) for t-AML patients compared to de novo AML, or secondary (from MDS) AML patients. There were an average of 10.2 ± 7.1 missense, nonsense, in-frame indel, or frameshift mutations per t-AML genome. Abnormalities of chromosomes 5 or 7 or complex cytogenetics were observed in 55 percent of the cases. The comparison de novo data were derived from 199 AML genomes or exomes,1 or 150 de novo MDS cases with extensive sequencing of candidate genes.2,3 For patients with t-AML, there was a higher frequency of mutations of TP53 (33% vs. 8%) and ABC (ATP-binding cassette) transporter proteins (that mediate cancer drug resistance), and a lower frequency of DNMT3A and NPM1 mutations, than in de novo AML. A multivariate analysis in 52 patients with t-AML revealed poor overall survival for those with KRAS or NRAS mutations (hazard ratio [HR], 5.33; p=0.002), IDH2 mutation (HR, 5.49; p=0.004), or TP53 mutation (HR, 3.52; p= 0.001).
For four of seven cases examined, the same TP53 mutation was present at low frequency (0.003% to 0.7%) in mobilized blood leukocytes or bone marrow three to six years before development of t-MDS/AML, and for two of the four patients, before any chemotherapy was administered. Development of the TP53 mutation preceded the del(5q) and del(7q) cytogenetic abnormalities that often occur in t-MDS and t-AML, or other driver mutations such as ETV6. By roughly five years from the initial detection of the abnormal clone to development of t-MDS or t-AML, TP53 mutation-bearing clones comprised 45 percent of the cells.
Given the lack of a significant increased number of mutations, implying that the issue of development of t-AML was not due to widespread DNA damage, the authors hypothesized that the TP53 mutated clone developed clonal dominance due to a competitive survival advantage. To prove this, the investigators used mixed bone marrow chimeras of wild-type and heterozygous mutant Tp53 (Tp53+/-) in mice to show that the Tp53+/- mutant clones indeed developed a competitive advantage after either chemotherapy or irradiation. These data are consistent with the notion that chemoradiotherapy leads to t-MDS and t-AML due to suppression of normal hematopoiesis and expansion of TP53-mutated clones.
In Brief
Recent publications have documented the development of age-related clonal hematopoiesis,4-6 including recurring AML mutations in a moderate proportion of healthy individuals. Dr. Wong and colleagues also examined healthy individuals aged 68 to 89 years and found that nine of 19 evaluable cases harbored TP53 mutations, with a variant allele frequency of 0.01 to 0.37 percent. Knowing that these mutations exist in healthy individuals, several questions arise for patients undergoing treatment with chemoradiotherapy: 1) Should these individuals be screened for TP53 and ABC transporter mutations? 2) What is the role of serial monitoring of mutant allele burden? 3) Could changes in the clonal architecture be used to reliably predict development of overt t-MDS/AML, and therefore, a basis for early intervention strategies? This work provides a more solid framework to start addressing some of these complex but important questions for our patients.
References
Competing Interests
Dr. Pamela Becker indicated no relevant conflicts of interest.