Based largely on vector innovation, gene therapy for hematologic disorders has advanced tremendously in the past decade, with 2014 witnessing sustained improvement in selected inherited diseases.
One example of how technological advances have salvaged the field is the successful therapy of X-linked severe combined immunodeficiency (Xl-SCID). This story arose from the ashes of the previously reported tragedy of insertional mutagenesis with the original γ-retroviral vectors, which led to acute leukemia in some patients due to activation of oncogenes including LMO2.1 Another γ-retroviral study this past year reported a substantial rate of development of acute leukemia among seven of 10 patients with Wiskott-Aldrich syndrome, in parallel with phenotypic correction of the defect in nine of 10 subjects.2 However, in a new, groundbreaking study, Dr. Salima Hacein-Bey-Abina and colleagues in Paris and in the United States utilized a novel safety-modified self-inactivating γ-retrovirus encoding the interleukin-2 receptor γ-chain (IL2Rγc) to transfer the normal gene to pediatric subjects.3 The Moloney murine leukemia virus LTR U3 enhancer was deleted from this modified vector, thereby reducing the mutagenic potential. Of the nine children with X1-SCID enrolled in the study, six exhibited sustained lymphocyte reconstitution including T and NK cells, with variable functional B-cell recovery. With a median of 33 months of follow-up, no leukemia had developed in any patient.
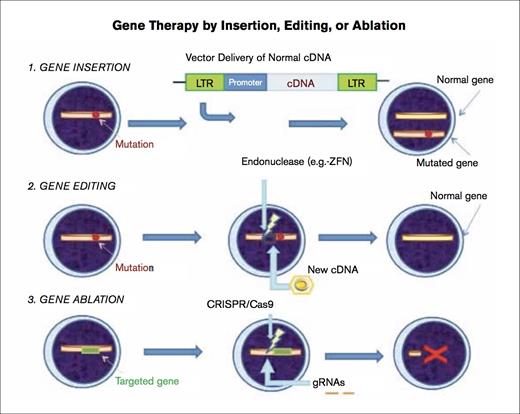
Gene Therapy by Insertion, Editing, or Ablation.1) Gene insertion – The mutation is depicted by the red spot. A vector (e.g., self-inactivating gammaretroviral or lentiviral vector) is used to introduce the normal cDNA encoding the entire protein into the cell under regulation by an appropriate promoter, and this vector integrates into the host cell genomic DNA. Transcription then proceeds, and the normal protein is produced. Both the mutated gene and the normal gene are present in the cell. 2) Gene editing – The affected cell shows the location of the mutation leading to the disorder with a red spot. A zinc finger nuclease (ZFN) is introduced, and cells are transduced with a vector, in this case, an integrase deficient lentiviral vector (IDLV) containing the normal partial cDNA for the affected gene, resulting in high-fidelity homology directed repair (HDR) and, thereby, correction of the defect. 3) Gene ablation – The CRISPR (clustered, regularly interspaced, palindromic repeats) –associated protein 9 (Cas9) nuclease is introduced in combination with short guide RNAs (gRNAs) that target the nuclease to the gene of interest (represented in green), resulting in cleavage of specific DNA sequence. Certain pairs of gRNAs successfully abrogate gene expression (depicted by red X).
Gene Therapy by Insertion, Editing, or Ablation.1) Gene insertion – The mutation is depicted by the red spot. A vector (e.g., self-inactivating gammaretroviral or lentiviral vector) is used to introduce the normal cDNA encoding the entire protein into the cell under regulation by an appropriate promoter, and this vector integrates into the host cell genomic DNA. Transcription then proceeds, and the normal protein is produced. Both the mutated gene and the normal gene are present in the cell. 2) Gene editing – The affected cell shows the location of the mutation leading to the disorder with a red spot. A zinc finger nuclease (ZFN) is introduced, and cells are transduced with a vector, in this case, an integrase deficient lentiviral vector (IDLV) containing the normal partial cDNA for the affected gene, resulting in high-fidelity homology directed repair (HDR) and, thereby, correction of the defect. 3) Gene ablation – The CRISPR (clustered, regularly interspaced, palindromic repeats) –associated protein 9 (Cas9) nuclease is introduced in combination with short guide RNAs (gRNAs) that target the nuclease to the gene of interest (represented in green), resulting in cleavage of specific DNA sequence. Certain pairs of gRNAs successfully abrogate gene expression (depicted by red X).
Another major advance of 2014 was the achievement by Dr. Amit Nathwani and colleagues in London and in the United States of long-term efficacy and safety of gene therapy for severe hemophilia B using a new vector, an adeno-associated virus serotype 8 (AAV8) vector, scAAV2/8-LP1-hFIXco, containing a codon-optimized modified factor IX transgene.4 The earlier intramuscular or intrahepatic studies using the AAV2 vector did not result in long-term expression of factor IX, and the intrahepatic study was associated with appreciable hepatic toxicity precluding that approach.5,6 But similar to the Xl-SCID study in which a novel vector design was critical to its success, a single intravenous infusion of this AAV8 vector resulted in an increase of circulating factor IX to a level of 1 to 6 percent of the normal value in all 10 patients over a median period of greater than three years. The most significant toxicity was mild elevation of the alanine amino-transferase level at seven to 10 weeks after infusion, which normalized after a tapering dose of prednisolone. Of the seven patients previously on factor replacement, four were able to discontinue, and the others were able to decrease the dose.
At the 2014 ASH Annual Meeting, two abstracts reported the use of a third type of vector, a lentivirus, to correct β-thalassemia.7,8 The replication-defective, self-inactivating lentiviral vector contains an engineered β-globin gene (βA-T87Q) and is called LentiGlobin BB305. Both studies reported that some study subjects no longer needed red blood cell transfusions.sevennineUnited States usedMoloney murine leukemia virus LTR U3 enhancer was deleted from this modified vector, thereby reducing the mutagenic potential. Of the nine children with X1-SCID enrolled in the study, six exhibited sustained lymphocyte reconstitution including T and NK cells, with variable functional B-cell recovery. With a median of 33 months of follow-up, no leukemia had developed in any patient.
2014 also witnessed innovation in preclinical studies in the methodology for gene editing, including not only the ability to correct in situ defective mutant genes, but also the ability to disrupt gene function. Gene editing is accomplished using endonucleases that can be targeted to specific regions of DNA to excise mutated region(s) and replace them with a normal sequence, or to disrupt genes to alter or eliminate undesirable gene function. The former technique was used to correct repopulating hematopoietic stem cells from a child with Xl-SCID.9 Dr. Fei Xie and colleagues from the University of California at San Francisco corrected induced pluripotent stem cells from patients with β-thalassemia using the endonuclease system CRISPR (clustered, regularly interspaced, palindromic repeats) associated protein 9 (Cas9) and the piggyBac cassette containing normal parts of the gene.10 The CRISPR/Cas9 endonuclease system was also used to disrupt two clinically relevant genes in primary human hematopoietic cells: 1) β-2 microglobulin as the accessory chain of the major histocompatibility gene class I molecules in order to produce hypoimmunogenic cells for transplantation, and 2) CCR5, the main co-receptor for certain strains of HIV that could potentially interfere with viral entry.11 These reports represent key milestones in gene editing technology that augur well for future success in the clinic.
References
Competing Interests
Dr. Becker indicated no relevant conflicts of interest.