Editor’s note
Data published in this paper were presented in abstract form by Dr. Kautz in the Plenary Session of the 2013 ASH Annual Meeting & Exposition in New Orleans.
After blood loss or hemolysis, erythropoiesis increases within a few hours and lasts for days in response to hypoxia-induced renal production of erythropoietin (EPO). A maximal erythropoietic response requires many factors, foremost of which is adequate iron for hemoglobin production. Studies of iron mobilization from cellular stores into the plasma and of duodenal iron absorption indicate that erythroblasts produce a factor(s) that contributes to the process that enhances iron availability in acute settings in which a maximal erythropoietic responses is needed.1 The hepatic hormone hepcidin, through its control of the cellular iron exporter, ferroportin, is the master regulator of iron homeostasis;2 and the absence of evidence for direct suppression of hepcidin by either EPO or hepatic hypoxia3 has suggested the existence of a hormone produced by erythroid cells that suppresses liver hepcidin expression to explain the observed increase in iron mobilization and absorption during periods of erythropoietic stress. In addition to being down-regulated by erythropoietic activity, hepcidin production, is up-regulated by inflammation and by intracellular iron content through the interleukin-6(IL-6)/JAK-STAT3 pathway and the bone morphogenetic protein(BMP)/SMAD pathway, repectively.2 By screening for mRNA expression of erythroid-specific secreted proteins induced rapidly after phlebotomy, Dr. Léon Kautz and colleagues in the laboratory of Dr. Tomas Ganz at the University of California, Los Angeles have identified an erythroid cell hormone, erythroferrone (ERFE) that suppresses hepcidin production through pathways other than IL-6/JAK-STAT3-BMP/SMAD and, thereby mediates an acute increase in iron mobilization and absorption during periods of erythropoietic stress (Figure)
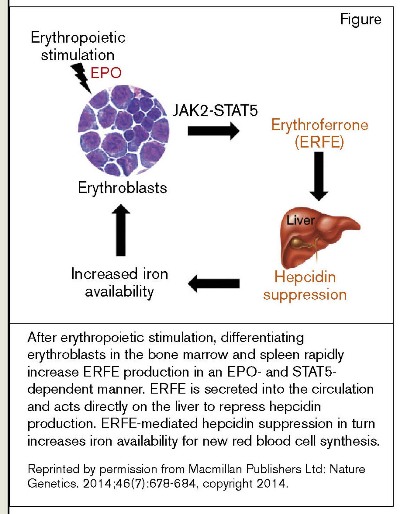
After erythropoietic stimulation, differentiating erythroblasts in the bone marrow and spleen rapidly increase ERFE production in an EPO- and STAT5- dependent manner. ERFE is secreted into the circulation and acts directly on the liver to repress hepcidin production. ERFE-mediated hepcidin suppression in turn increases iron availability for new red blood cell synthesis.Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics. 2014;46(7):678-684, copyright 2014.
After erythropoietic stimulation, differentiating erythroblasts in the bone marrow and spleen rapidly increase ERFE production in an EPO- and STAT5- dependent manner. ERFE is secreted into the circulation and acts directly on the liver to repress hepcidin production. ERFE-mediated hepcidin suppression in turn increases iron availability for new red blood cell synthesis.Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics. 2014;46(7):678-684, copyright 2014.
Kautz et al. demonstrated that many tissues express ERFE, but within four hours of phlebotomy or EPO administration, expression increased in cells of all stages of erythroid differential such that the bone marrow became the major source of ERFE production. Inhibition of the Jak2-Stat5 signaling mechanism downstream of the EPO receptor greatly diminished ERFE induction. ERFE-deficient mice have normal steady-state hemoglobin values as adults, but they have delayed hemoglobin recovery after phlebotomy and lower than normal hemoglobin values during the period of expanded erythropoiesis that accompanies rapid growth at six weeks of age. The normal suppression of hepatic hepcidin expression following phlebotomy was essentially absent in ERFE-deficient mice. Administration of recombinant ERFE or chronic expression of ERFE in mice transduced with ERFE cDNA suppressed hepcidin production.
In Brief
The identification of ERFE as an erythropoietic regulator of iron mobilization and absorption through hepcidin suppression has scientific and clinical implications. The hepatic ERFE receptor and its intracellular signaling mechanisms are unknown, but a candidate transcription factor, ATOH8, was first associated with iron metabolism by Kautz et al.4 ATOH8 regulates transcription of the gene-encoding hepcidin through a mechanism separate from the BMP/SMAD pathway, and hepatic ATOH8 levels are reduced during erythropoietic stress.5 In a broader context, ERFE is expressed in skeletal muscle where it is recognized as a C1q/TNF-related protein (CTRP), termed myonectin (CTRP 15), which regulates lipid metabolism.6 Myonectin, which is induced after fasting by re-feeding or nutrient administration, acts through PI3-K/akt/mTOR signaling to inhibit hepatocyte autophagy,7 suggesting a possible role for this pathway in hepcidin regulation. In clinical hematology, excess iron accumulation complicates diseases with a major component of ineffective erythropoiesis, such as β-thalassemia, in which erythroblast numbers are greatly expanded, but the erythroblasts undergo intramedullary apoptosis before completing differentiation.8 Greatly expanded erythroblasts in ineffective erythropoiesis produce large amounts of ERFE, and treatment that mitigates ERFE effects would be expected to reduce inappropriate iron absorption. Indeed, using a model of β-thalassemia intermedia in which mice also have ERFE gene knockout, Kautz and colleagues showed that serum hepcidin is normalized and hepatic non-heme iron content is lower while Hgb values are similar when compared with thalassemic mice with normal ERFE activity. Conversely, increased hepcidin in the anemia of chronic inflammation restricts the erythropoietic response by limiting the availability of iron. Development of a therapy that stimulates ERFE production or activity could potentially decrease hepcidin in this setting and, thereby ameliorate the iron-restrictive component of the anemia.
References
Competing Interests
Dr. Koury indicated no relevant conflicts of interest.