Much progress has been made in the quest to conquer, through gene therapy, monogenetic diseases rooted in the hematopoietic stem cell (HSC), but challenges remain. Until recently, methods to “correct” the genetic defect involved introducing into the HSC genome a wild-type copy of the mutant gene, using a retroviral vector-based system. This technique has yielded considerable success, but concerns about insertional mutagenesis and difficulty procuring sufficient HSCs for transduction, preserving transduced HSCs in an undifferentiated state ex vivo with intact engraftment capability have pushed investigators to identify techniques to enhance both safety and efficacy. One area where recent progress is notable is development of targeted DNA editing techniques that allow true correction of genetic defects in situ. Such editing functions use endonucleases, including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases, or RNA-guided nucleases (e.g., the clustered, regularly interspaced, short palindromic repeat [CRISPR]-associated system [Cas]). Because only a small amount of DNA that encompasses the mutated site is excised by the endonuclease and repaired by naturally occurring mechanisms in situ, these techniques have the advantage over retroviral vector-based gene therapy in that native gene regulatory elements (e.g., promoters, enhancers) are preserved. The other major advantage of targeted editing over retroviral-based gene therapy is that the potential toxicity associated with random integration of the retrovirus (i.e., activation of oncogenes that can lead to development of acute leukemia, as reported in the early clinical trials) is likely eliminated.
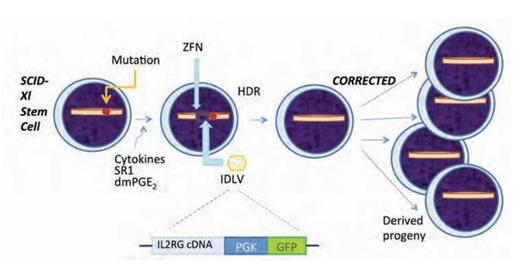
Now, using a ZFN-based technique, Dr. Pietro Genovese and colleagues at the San Raffaele Telethon Institute for Gene Therapy in Milan, Italy, have successfully corrected, in long-term repopulating HSCs, the disease-causing mutation in the interleukin-2 receptor gamma (IL2RG) chain in a patient with X-linked severe combined immunodeficiency (SCID-Xl). Two major innovations were critical for this accomplishment: 1) refinement of the technique to induce cycling of HSCs without differentiation so as to take advantage of high-fidelity homology-directed DNA repair (HDR); and 2) development of the method to efficiently introduce into cells both the nuclease and the cDNA template needed to correct the mutant gene (Figure). Umbilical cord blood stem/progenitor cells were incubated with the cytokines stem cell factor, Flt3 ligand, thrombopoietin, and interleukin-6, together with StemReginin 1 and 16,16-dimethyl-prostaglandin E2, such that cells would progress through the cell cycle (a requirement for HDR) but not undergo differentiation. For correction of the SCID-Xl mutation, the integrase-deficient lentiviral vector (IDLV) contained a cDNA construct composed of exons 5-8 of the IL2RG gene and the green fluorescent protein (GFP) marker (Figure). The ZFN was designed to excise exon 5 of IL2RG, and when the excised site was repaired by HDR using the cDNA construct contained in the IDLV as the template, the mutated DNA was repaired. Thus, the cDNA construct developed by Dr. Genovese et al. can be used to correct the genetic defect in any SCID-X1 case that arises from a mutation affecting any nucleotide residues downstream of exon 4. Analysis of the treated cells showed expression of the GFP marker gene by 100 percent of the primitive stem cells when transduction was induced on the third day of culture. The progeny of the corrected HSCs included myeloid and erythroid cells, as well as T and NK cells. The latter two cell lineages are strictly dependent on normal function of the IL2RG gene, and expression of the repaired gene was demonstrated by massive expansion of GFP marked T and NK cells that were shown to have the capacity to mediate rejection of an allogeneic tumor cell line.
Correction of X-linked Severe Combined Immunodeficiency (SCID-X1) by Gene Editing. The illustration of the stem/progenitor cell on the far left depicts the location of the gene mutation that causes the disorder. The affected cells are incubated in a cytokine cocktail that induces cell cycling without differentiation. Next, the targeted zinc finger nuclease (ZFN) is introduced into the cell by electroporation, followed by transduction with integrase deficient lentiviral vector (IDLV). The IDLV contains the cDNA for IL2RG exons 5-8 and green fluorescent protein (GFP) driven by the phosphoglycerate kinase (PGK) promoter. GFP functions as an easily identifiable marker expressed in parallel with IL2RG. This process results in high-fidelity homology directed repair (HDR), and thereby, corrects the gene mutation that caused SCID-X1. The corrected stem cells can be used therapeutically as, following transplant, their hematopoietic progeny, including T cells and NK cells, will expand in vivo and thereby restore normal immune function (far right).
Correction of X-linked Severe Combined Immunodeficiency (SCID-X1) by Gene Editing. The illustration of the stem/progenitor cell on the far left depicts the location of the gene mutation that causes the disorder. The affected cells are incubated in a cytokine cocktail that induces cell cycling without differentiation. Next, the targeted zinc finger nuclease (ZFN) is introduced into the cell by electroporation, followed by transduction with integrase deficient lentiviral vector (IDLV). The IDLV contains the cDNA for IL2RG exons 5-8 and green fluorescent protein (GFP) driven by the phosphoglycerate kinase (PGK) promoter. GFP functions as an easily identifiable marker expressed in parallel with IL2RG. This process results in high-fidelity homology directed repair (HDR), and thereby, corrects the gene mutation that caused SCID-X1. The corrected stem cells can be used therapeutically as, following transplant, their hematopoietic progeny, including T cells and NK cells, will expand in vivo and thereby restore normal immune function (far right).
Using this same technique, edited HSCs derived from normal adult bone marrow were shown to engraft in a mouse xenograft model, and cells from a four-month-old SCID-Xl patient with a missense mutation in exon 7 were corrected using this novel methodology.
In Brief
SCID-Xl was among the first diseases in which gene therapy was successful using defective retroviruses to transfer a normal copy of the cDNA into mutant cells.1,2 Although this technique was effective in restoring T-cell immunity to the subjects, the procedure was associated with the risk of random insertion of the genetic material into the genome that resulted in development of acute leukemia through activation of oncogenes such as LMO2. Fortunately, four out of five of the children who developed acute leukemia due to insertional mutagenesis were successfully treated with chemotherapy. The elegant studies of Dr. Genovese and colleagues demonstrate proof of principle that gene editing can be incorporated into gene therapy strategies for disorders involving the HSC. Whether this new approach is clinically viable remains to be proven, and the long-term consequences of this procedure including persistence of functional correction and degree of toxicity must be evaluated in clinical trials.
References
Competing Interests
Dr. Becker indicated no relevant conflicts of interest.