Therapeutic Targeting of Canonical NF-κB Pathway in MLL-Fusion Leukemia. Reprinted from Cancer Cell, Vol. 24. Susmu Goyama, James C. Mulloy. NF-κB: A Coordinator for Epigenetic Regulation by MLL. Pages 401-402. Copyright 2013, with permission from Elsevier.
Therapeutic Targeting of Canonical NF-κB Pathway in MLL-Fusion Leukemia. Reprinted from Cancer Cell, Vol. 24. Susmu Goyama, James C. Mulloy. NF-κB: A Coordinator for Epigenetic Regulation by MLL. Pages 401-402. Copyright 2013, with permission from Elsevier.
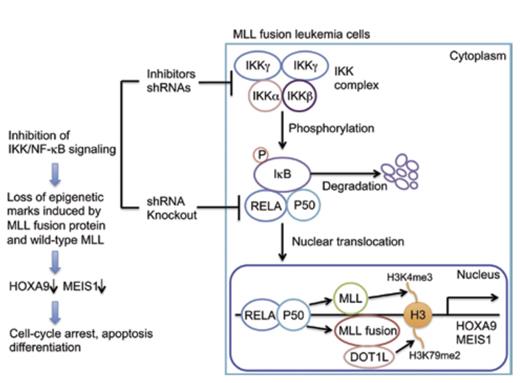
The MLL gene is located at chromosome 11q23 and encodes a protein with epigenetic activity. The product of the gene is an enzyme that modifies chromatin by methylating lysine 4 on histone H3 (Figure). The SET domain of MLL that bestows this methyl transferase activity is deleted by the chromosomal translocations that occur in MLL-rearranged acute leukemias, and consequently, aberrant epigenetic functions that contribute to disease pathophysiology are acquired (Figure). MLL-rearrangement is observed in subsets of both myeloid and lymphoid acute leukemias that are characterized by an aggressive clinical course and a poor prognosis.
In the context of acute leukemia, MLL is promiscuous in that rearrangements involve a variety of partner genes that generate leukemogenic fusion proteins with various functional properties that mediate disease by driving proliferation, enhancing survival, or inducing differentiation arrest. At least 70 translocations involving MLL have been documented, including those that partner with lens epithelium-derived growth factor; the histone methyltransferase, DOT1L (Figure); chromobox homolog 8; histone demethylase KDM1A; and bromodomain-containing 4. These fusion proteins activate key regulatory pathways that contribute to leukemia pathogenesis (Figure). With so many different fusion partners, it seemed unlikely that the pathobiologic process underlying leukemia involving MLL rearrangement would converge on a common mechanism, but now, Kuo and colleagues report that activation of the IKK/NFĸB pathway is a critical component of the neoplastic process in MLL-rearranged leukemias (Figure).
The transcription factor NFĸB has a fundamental role in cancer pathogenesis, through its effects on cell proliferation, its anti-apoptotic properties, and its contributions to drug resistance and metastasis. The transcription factor complex has two forms: NFKB1, which drives the canonical pathway and consists of RELA/p50; and NFKB2, which drives the non-canonical pathway and consists of RELB/p52. The canonical pathway affects cell proliferation, survival, and self-renewal (Figure). The activity of NFĸB is tightly controlled by an inhibitory protein (IkB) that binds and sequesters the canonical RELA/p50 complex in the cytoplasm (Figure). NFĸB becomes active when IkB is phosphorylated by the IKK complex (Figure). Consequently, IkB is degraded and RELA/p50 is free to translocate to the nucleus and enable transactivation of target genes (Figure). For example, transcriptional activation of two key target genes, HOXA9 and MEIS1, by NFKB1 contributes to the self-renewal capacity of leukemia stem cells (Figure).
To identify proteins that underlie neoplastic transformation in MLL-rearranged leukemia, a lentiviral-based, shRNA-knockdown approach was used by Kuo and colleagues to examine the effect of 211 candidate stem cell kinases or phosphatases on the growth of MLL-AF9 transduced cells. Knockdown of expression of 33 genes resulted in > 50 percent impairment of growth. Notably, several of the genes identified in these experiments, including IKKα, IKKβ, IKKγ, PLK1, PPP4C, and IRAK3, are components of the NFĸB pathway. In addition, the authors observed that AML cells with MLL translocation exhibit hypomethylation of genes that organize into NFĸB networks. In supportive experiments, inhibitors of the IKK complex were shown to selectively retard growth of MLL-rearranged leukemia cells, and knockdown of the NFĸB subunit RELA impaired growth of MLL cell lines MV4;11 and ML-2, but not of K562 cells, which do not have an MLL translocation (Figure). In a murine model of MLL leukemogenesis, IKK inhibitors or knockdown of either IKK complex components or RELA abrogated the leukemogenicity of MLL oncogene-transduced cells, and RELA overexpression in MLL-AF6-transduced cells shortened both latency of development of leukemia and survival. RELA knockdown, in both MLL-transduced mouse progenitors and human leukemia cell lines, also markedly reduced HOXA9 and MEIS1 transcript levels, whereas forced RELA expression increased these levels. Additional in vitro experiments demonstrated that IKK inhibition decreases proliferation, increases apoptosis, and induces differentiation in MLL-AF10 murine leukemia cells and that IKK inhibition markedly downregulates expression of genes associated with poor prognosis in AML while simultaneously upregulating those associated with a good prognosis.
In Brief
Together, these rigorous studies demonstrate that NFκB signaling is critical for propagation and maintenance of MLL-associated leukemias and for implementation of the epigenetic programs directed by the MLL fusion proteins. This enlightening publication highlights NFĸB as the final common pathway for the MLL-rearranged leukemias and, in so doing, reveals new targets for therapy of these aggressive, often refractory, hematologic malignancies. As an example, a selective amino-nucleoside inhibitor, EPZ-5676, which targets the DOT1L complex consisting of DOT1L, a histone methyltransferase (Figure), and MLL fusion partners such as AF9, ENL, and AF10, has been studied in preclinical models,1 and a phase I clinical trial using EPZ-5676 is ongoing.
References
Competing Interests
Dr. Becker indicated no relevant conflicts of interest.