Senior Member, H. Lee Moffitt Cancer Center, and Professor, University of South Florida, Tampa, FL
Dr. Epling-Burnette indicated no relevant conflicts of interest.
Large granular lymphocyte (LGL) leukemia is a rare (but probably under-diagnosed) chronic lymphoproliferative neoplasm that shares overlapping genetic and clinical characteristics with Felty syndrome.1 Unlike most lymphoproliferative processes that produce symptoms due to either clonal expansion within the bone marrow or infiltration by the neoplastic cells of lymphoid and/or non-lymphoid organs, the clinical manifestations of LGL leukemia (cytopenias [particularly neutropenia] and rheumatic and constitutional symptoms) are a consequence of immune dysregulation and aberrant cytokine production.1 First described in 1977 by McKenna et al.2 and shown to be of clonal origin in 1985 by Loughran et al.,3 the disease was identified based on the presence of morphologically atypical lymphocytes with prominent azurophilic cytoplasmic granules. Flow cytometric analysis identified these cells as mature CD8+ T cells that co-expressed CD16 (IgG Fc receptor III). A clinically similar disorder was later described in which the LGL cells had a natural killer cell phenotype (CD3-, CD16+, CD56+). While largely thought to develop as a consequence of a sustained immune reactive process due to chronic antigen stimulation, identification of T-cell receptor (TCR) gene rearrangement in the T-cell variant indicated a clonal origin and infiltration by the abnormal T- or NK-cells in the liver, lung, and spleen led to inclusion by the World Health Organization (WHO) of the disease among lymphoid malignancies.3 (The WHO classification is based on the affected lineage, defined as either chronic lymphoproliferative disorders of NK cells [CLPD-NKs] or T-cell large granular lymphocytic leukemia [T-LGL]).
A prescient 2001 study showed that STAT3 was activated in peripheral blood mononuclear cells from patients with T-cell LGL leukemia,4 but the molecular basis of this clinically heterogeneous but morphologically distinct entity remained enigmatic until recently when an international group of investigators identified somatic mutations in the signal transducer and activator of transcription 3 (STAT3) gene in 31 of 77 cases (40%).5 A subsequent paper extended the initial studies, reporting a mutation frequency of 28 percent (48 out of 170 cases) and documented a similar mutational rate in both the NK- and T-cell variants.6 Identification of this somatic mutation confirms the clonal nature of both variants of LGL leukemia and provides new insights into the molecular mechanisms that underlie T-cell and NK-cell proliferation.
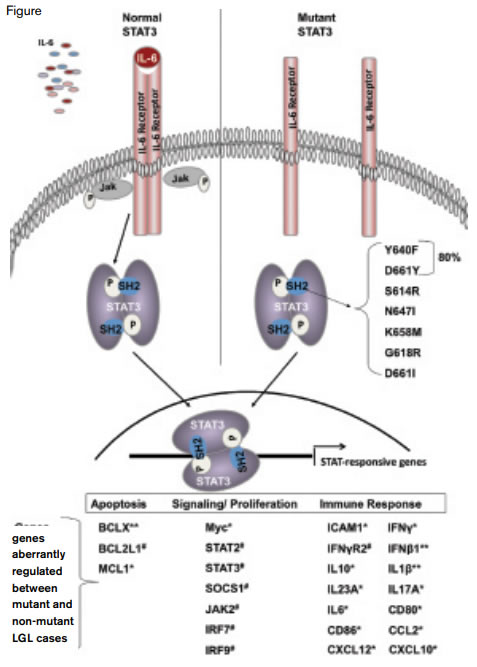
The STAT3 Pathway.Left, normally, binding of IL-6 to its cognate receptor activates the JAK pathway that in turn induces homodimerization of STAT3. The STAT3 homodimer migrates to the nucleus where it initiates transcription of a variety of genes involved in apoptosis, signaling, and proliferation and the immune response.
The STAT3 Pathway.Left, normally, binding of IL-6 to its cognate receptor activates the JAK pathway that in turn induces homodimerization of STAT3. The STAT3 homodimer migrates to the nucleus where it initiates transcription of a variety of genes involved in apoptosis, signaling, and proliferation and the immune response.
STAT3, a cytoplasmic transcription factor that translocates to the nucleus, was originally identified as an acute phase response factor and a central component of the downstream molecular pathway that mediates signaling induced by the IL-6-family of cytokines that share a common gp130 receptor subunit that initiates the signaling cascade (Figure). Binding of ligand induces receptor dimerization that leads to activation of members of the Janus kinase family (Jak1, Jak2, Jak3, and Tyk2), which, in turn, causes STAT protein dimerization, nuclear translocation, DNA binding, and transcriptional activation of target genes (Figure). The somatic mutations identified by both Koskela et al.5 and Jerez et al.6 reside within the Src homology 2 (SH2) phosphotyrosine-binding domain of STAT3 at the dimerization interface (Figure). Of 49 STAT3 mutations identified by Jerez and colleagues,6 80 percent were either Y640F or D661Y (Figure). These mutations result in amino acid substitution in the major protein-protein interaction domain in which an interface is formed with the carboxyterminal phospho-tyrosine 705 (Y705) that is responsible for homo and heterodimerization. Although the specific consequence of the mutation is undefined, constitutive STAT3 DNA binding,4 increased expression of STAT3-responsive genes,5,6 and in vitro functional studies showing both increased transcriptional activity and TAT3-inhibitor-responsive death suggest that STAT3 mutations in LGL leukemia lead to a gain-of-function phenotype that contributes to survival of the leukemic LGL cells.
The first linkage between STAT3 signaling and T-cell leukemia pathogenesis was based on studies of HTLV-1-mediated adult T-cell leukemia (ATL) where constitutive phosphorylation of JAK proteins was shown to mediate tumor growth by HTLV-1.7 Studies in mice and humans have defined crucial roles for STAT3 in cytokine signaling, embryogenesis, and malignant transformation.8 Targeted deletion of Stat3 using the Cre-loxP recombination system demonstrated the importance of this transcription factor in T-cell cytokine signaling.9 Thymocytes from Lck-Cre/Stat3flox/- mice maintained normal function in response to STAT5-specific cytokines IL-2 and IL-7, but a markedly reduced proliferative response to STAT3-specific IL-6 stimulation. STAT3-deficient T-cells were resistant to IL-6 induced antiapoptotic responses, which is consistent with the proposed role of this molecule in LGL leukemia. In the case of LGL leukemia, STAT3 activation in patients with or without the mutation is associated with an anti-apoptotic phenotype.4-6
Discovery of somatic mutations of STAT3 has provided new insights into the molecular basis of LGL leukemia, established a platform for creating novel diagnostic tools, and suggested the possibility of developing a targeted approach to therapy. Patients with STAT3 mutations had earlier onset of symptomatic disease, a history of multiple treatments, and were more likely to have neutropenia and rheumatoid arthritis compared with patients without STAT3 mutation. Conceivably, mutant STAT3 may be involved in the pathobiology of other disease processes such as Felty syndrome, aplastic anemia, and myelodysplastic syndrome where LGL expansion is observed. Furthermore, it seems likely that additional genetic mutations, aberrant epigenetic processes, or microenvironmental abnormalities will be identified that regulate STAT3 in LGL cells because functional studies demonstrate aberrant JAK/STAT3 signaling in the absence of STAT3 mutations (Figure.).4-6 Further studies will be required to confirm these initial observations and to determine the pathophysiologic basis of the difference in clinical phenotype. Together with observations in myeloproliferative neoplasms, the current studies underscore the importance of JAK/STAT pathways in the molecular pathogenesis of hematologic neoplasms.