Clonal hematopoiesis (CH) is the name given to the emergence of somatically mutated hematopoietic stem cell clones detectable by next-generation sequencing of circulating leukocytes or bone marrow cells. The condition is a common consequence of aging and, depending on the threshold of mutated DNA used (typically 2%), affects up to 30 percent of patients older than 80 years.1,2 The expansion of CH clones appears to relate to their survival advantage in the marrow microenvironment relative to normal stem cells and is facilitated by cytotoxic chemotherapy and radiation therapy.3,4 Most individuals with CH have normal blood counts. When this is the case, the term “CH of indeterminate potential” or “age-related CH” (ARCH) is used; however, when CH is identified in patients who have been treated for cancer with cytotoxic therapies, they are associated with a risk of subsequent development of myeloid neoplasms, including therapy-related myeloid neoplasms (t-MNs).5,6 In the cancer survivor, therapy-related myelodysplastic syndrome/acute myeloid leukemia is a devastating complication associated with dismal prognosis. While the association between CH identified after cancer therapy and t-MN is clear, the majority of patients with CH (even involving high-risk mutations such as TP53) do not evolve to t-MN following therapy.6,7 Thus, there is a need to balance the benefit derived from chemotherapy and radiation therapy with the risk of a subsequent aggressive hematologic malignancy. Patients would benefit from a more precise and accurate quantification of this risk and a deeper understanding of factors that contribute to the progression from CH to t-MN.
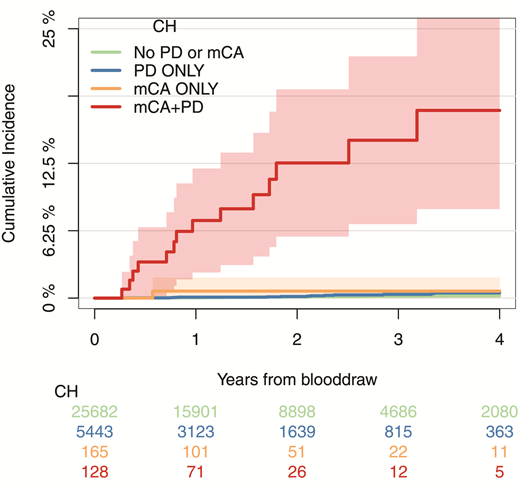
Dr. Teng Gao and colleagues recently analyzed the blood samples of 32,422 patients with cancer, including a large subset previously exposed to cytotoxic cancer therapies, for somatic mutations in 468 genes and for mosaic chromosomal alterations (mCAs; representing amplifications, deletions, or loss-of-heterozygosity of genetic material), using probes against common single-nucleotide polymorphisms distributed across the genome. As expected, they identified somatic mutations representing CH in approximately one-third of the patients (ranging from 5% of patients in their 20s to 60% of patients in their 80s). However, they also identified mCAs in 346 individuals, which paralleled the age-associated increase in CH (mCAs ranged from <1% in patients <50 years to >3% in patients >80 years). Although patients with mCAs had blood counts similar to those of patients lacking mCAs, the presence of mCAs was significantly associated with the presence of somatic mutations, particularly with high numbers of mutated genes at high variant allele fractions. Interestingly, the latter features have been associated with high risk of progression of CH in cytopenic patients to myeloid neoplasia.8 Moreover, the mCAs often involved the same sites bearing gene mutations, suggesting a synergistic relationship. For example, loss of heterozygosity at 7q was associated with mutations in EZH2, a gene located at the 7q36 locus. During the follow-up period of three years after the genetic testing, 14.6 percent of patients who had combined mCAs and somatic mutations developed a hematologic malignancy, compared to less than 1 percent of patients with mCAs alone, somatic mutations alone, or no genetic abnormalities (Figure). In multivariable analysis, the effect of mCA on the development of subsequent hematologic malignancy was independent of the effect of the number of somatic mutations and their variant allele frequencies.
Cumulative incidence of leukemia (including myelodysplastic syndromes, myeloproliferative neoplasms, acute myeloid leukemia, chronic myeloid leukemia, and chronic lymphocytic leukemia) among patients who had detectable mosaic chromosomal alterations (mCA) only, gene mutation putative driver (PD) only, both, or neither; 95% CIs are shown in shaded ribbons. From Figure 4c. Gao T, Ptashkin R, Bolton KL et al. Nat Commun 2021;12:338. Licensed under Creative Commons Attribution 4.0 International License; http://creativecommons.org/licenses/by/4.0/.
Cumulative incidence of leukemia (including myelodysplastic syndromes, myeloproliferative neoplasms, acute myeloid leukemia, chronic myeloid leukemia, and chronic lymphocytic leukemia) among patients who had detectable mosaic chromosomal alterations (mCA) only, gene mutation putative driver (PD) only, both, or neither; 95% CIs are shown in shaded ribbons. From Figure 4c. Gao T, Ptashkin R, Bolton KL et al. Nat Commun 2021;12:338. Licensed under Creative Commons Attribution 4.0 International License; http://creativecommons.org/licenses/by/4.0/.
In Brief
The study by Dr. Gao and colleagues provides a glimpse into the initial steps in progression from a state of CH to myeloid malignancy: Gene copy-number alterations engendered by mCAs cooperate with somatic mutations to drive this process. Chromosomal copy-number abnormalities have also been shown to correlate with progression in other hematologic neoplasms, such as monoclonal gammopathy of undetermined significance and monoclonal B-cell lymphocytosis.9,10 Additionally, the study provides a critical tool to inform risk-benefit analysis for patients with cancer who are contemplating adjuvant therapy. Patients with CH are indeed at increased risk of developing t-MN, but the subset of patients with mCAs represents those with the highest risk. It is the latter group of patients in whom physicians may consider eliminating or reducing adjuvant therapy in an effort to mitigate the risk of developing a subsequent hematologic malignancy. The number and type of mutations and their variant allele frequencies may still factor into risk assessment of patients with CH, but mCA represents a powerful new indicator that seems to be superior to these ”traditional” methods of evaluating the risk of CH progression. While the SNP array used to detect mCAs in this study was not as sensitive as the next-generation sequencing assay for somatic mutations (≥10% vs. ≥2%, respectively), this technique can be applied to peripheral blood samples and can detect small regions of alteration as well as copy-neutral loss of heterozygosity, unlike conventional cytogenetics and fluorescence in situ hybridization. The study also raises several interesting questions that warrant further study: Do mCAs also contribute to the increased risk of cardiovascular mortality associated with CH of indeterminate potential? Does the synergistic effect of mCAs on somatic mutations extend to CH in the noncancer patient setting and to CH after treatment for acute myeloid leukemia? Will interventions that may abrogate the risk of CH progression to myeloid neoplasia still be effective if the somatic mutations are accompanied by mCAs? By diversifying our interrogation of hematopoietic clones to include chromosomal abnormalities, we will have the opportunity to better understand the early development of myeloid neoplasia and better counsel and manage patients in whom such clones are detected.
Competing Interests
Dr. Hasserjian indicated no relevant conflicts of interest.