Abstract
Philadelphia chromosome-like acute lymphoblastic leukemia (Ph-like ALL) is a common subtype of B-lineage acute lymphoblastic leukemia (B-ALL) with increasing frequency across the age spectrum. Characterized by a kinase-activated gene expression profile and driven by a variety of genetic alterations involving cytokine receptors and kinases, Ph-like ALL is associated with high rates of residual disease and relapse in patients treated with conventional chemotherapy. In this case-based review, we describe the biology of the 2 major ABL-class and JAK pathway genetic subtypes of Ph-like ALL, discuss current diagnostic testing methodologies, and highlight targeted inhibitor and chemo/immunotherapy approaches under clinical investigation in children, adolescents, and adults with these high-risk leukemias.
Learning Objectives
Understand the spectrum of clinical tests available for the screening and diagnosis of Philadelphia chromosome-like acute lymphoblastic leukemia (Ph-like ALL)
Recognize indications for incorporating tyrosine kinase inhibitors into the treatment of patients with Ph-like ALL
Biology of Philadelphia chromosome-like acute lymphoblastic leukemia
Philadelphia chromosome-like acute lymphoblastic leukemia (Ph-like ALL) is a subset of B-lineage acute lymphoblastic leukemia (B-ALL) associated with high relapse risk and inferior outcomes in children and adults despite modern risk-adapted chemotherapy regimens.1-8 Originally discovered in 2009 via gene expression profiling studies of cytogenetically normal National Cancer Institute (NCI) high-risk (HR) B-ALL cases, researchers from the Children's Oncology Group (COG)/St Jude Children's Research Hospital/University of New Mexico and from the Dutch Children's Oncology Group independently identified a group of patients with a gene expression signature similar to those with Ph-positive (Ph+) ALL but without the canonical BCR-ABL1 fusion, leading to its BCR-ABL1–like ALL or Ph-like ALL nomenclature.9,10 The Ph-like subtype accounts for 5% to 10% of NCI standard-risk11 and 15% to 20% of NCI HR childhood B-ALL,3 as well as 25% to 30% of young adults with B-ALL aged 21 to 39 years.1 Ph-like ALL was recognized as a provisional disease entity in the World Health Organization 2016 classification of acute leukemias.12 Over the past decade, the advent of RNA- and DNA-based next- generation sequencing (NGS) technologies has deepened our understanding of the genomic landscape of Ph-like ALL, which is characterized by a diverse spectrum of kinase-activating alterations likely amenable to molecularly targeted therapies, as evidenced by a vast body of preclinical data and increasing clinical evidence.1,13-17
The 2 most clinically relevant Ph-like ALL subgroups are those with (1) JAK-STAT pathway alterations and (2) ABL- class gene rearrangements. JAK-STAT pathway alterations comprise approximately 70% of Ph-like ALL cases and can be further subdivided by their specific kinase-activating lesions: (a) CRLF2 rearrangements with 5′ partners P2RY8 or IGH account for 50% of Ph-like ALL, and half of CRLF2-rearranged cases harbor concomitant JAK2 or JAK1 point mutations; (b) JAK2 or EPOR fusions collectively comprise 10% to 15% of Ph-like ALL; and (c) sequence mutations involving IL7R, SH2B3, IL2RB, or TYK2 genes occur in a smaller number of cases.1,3,18 The second major Ph-like molecular subgroup involves rearrangements in ABL-class family genes, including ABL1, ABL2, CSF1R, PDGFRA, and PDGFRB, that collectively comprise approximately 15% of Ph-like ALLs.1,3,18
The heterogeneity of kinase-activating alterations in Ph-like ALL, most of which are cryptic by conventional cytogenetics, renders its diagnosis complex in clinical practice and ideally requires access to specialty NGS testing. As several precision medicine trials are actively screening patients' diagnostic ALL specimens for Ph-like genetic alterations and prospectively evaluating the potential efficacy of tyrosine kinase inhibitor (TKI) addition to conventional multiagent chemotherapy, timely and accurate identification of these high-risk patients to guide trial assignment and/or tailor targeted therapies is essential. This review focuses upon description of modern clinical diagnostics for Ph-like ALL and highlights the evolving role of TKI-based therapy for patients with CRLF2/JAK pathway or ABL-class genetic alterations.
CLINICAL CASE
A 6-year-old white boy was diagnosed with NCI HR B-ALL with a presenting white blood cell count of 500 000/µL with 92% peripheral blasts and microscopic leukemia involvement of his central nervous system. He was started on a 4-drug induction therapy (vincristine, daunomycin, peg-asparaginase, and dexamethasone) with intrathecal methotrexate chemotherapy on the COG trial AALL1732 phase 3 clinical trial (NCT03959085) and was observed to have slow reduction of leukocytosis during the first week of chemotherapy. Institutional B-ALL fluorescence in situ hybridization (FISH) testing was negative for chromosomal trisomies and intrachromosomal amplification of chromosome 21, ETV6-RUNX1, BCR-ABL1, TCF3, and KMT2A rearrangements. Low-density microarray testing returned positive for the Ph-like gene signature with low CRLF2 expression. Extended ABL-class FISH testing using ABL1, ABL2, and CSF1R/PDGFRB break-apart FISH probes was positive for a PDGFRB gene rearrangement. The patient was removed from the AALL1732 study at day 8 of induction for addition of imatinib 340 mg/m2 daily to chemotherapy. RNA-based Archer FusionPlex and DNA-based NGS analyses subsequently confirmed EBF1-PDGFRB fusion and IKZF1 and CDKN2A deletions. The child achieved morphologic remission at end of induction but had positive measurable/minimal residual disease (MRD) of 4.9%. His family consented to postinduction chemotherapy on the European Study of Postinduction Treatment of Ph-Positive ALL (EsPhALL) 2017/COG AALL1631 trial (NCT03007147) with imatinib addition, which decreased his MRD to2 × 10−4 (0.02%) by immunoglobulin/T-cell receptor polymerase chain reaction assessment at the end of induction 1B. He was stratified as standard risk given this favorable MRD response and was eligible for randomization between arm A (EsPhALL backbone) vs arm B (COG HR ALL backbone) chemotherapy. There was no indication for hematopoietic stem cell transplantation (HSCT) in first complete remission (CR1). He was assigned to arm B and had continuous imatinib with postinduction chemotherapy as per COG AALL1131 or AALL1732 arm A for a planned 2-year total treatment duration.
Clinical testing modalities for the diagnosis of Ph-like ALL
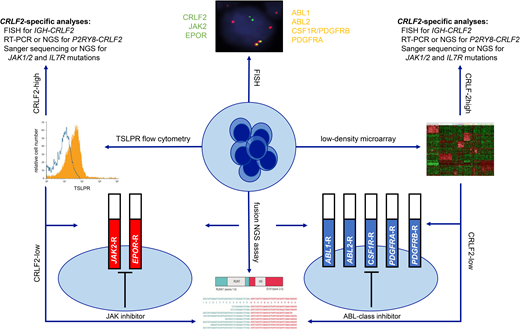
The heterogeneous spectrum of kinase-activating alterations, cryptic nature of these genetic aberrations on conventional cytogenetics, and complexity of testing to date have rendered Ph-like ALL a diagnostic challenge in clinical laboratories. Two main RNA-based strategies are presently used to screen for Ph-like ALL, which involve assessment of the pathognomonic kinase-activated gene signature via microarray analysis and/or direct detection of Ph-like kinase fusions (Table 1). Gene expression profiling, the initial discovery modality used in European and North American studies,9,10 is not available in most clinical laboratories and has been replaced by a TaqMan low-density microarray (LDA) microfluidic card, which measures the expression of a panel of 8 or 15 genes to identify the Ph-like gene signature.19 Gene expression of interest is normalized to a housekeeping gene to derive an integrated score between 0 and 1, with scores ≥0.5 considered positive for the Ph-like signature. Higher LDA scores >0.7 are usually indicative of an ABL-class, JAK2, or EPOR Ph-like kinase fusion, while CRLF2 rearrangements occur across a broader range of LDA positivities.19 The COG has employed LDA screening as a rapid (~48-hour turnaround time) and cost- effective modality to identify the 15% to 25% of patients with NCI HR ALL and the Ph-like gene signature who require further molecular testing to identify their underlying kinase-activating alterations. The LDA card also enables direct detection of P2RY8-CRLF2 fusions and quantification of CRLF2 and EPOR expression levels that, when elevated, may predict true genetic rearrangements usually identifiable by FISH or RNA-based fusion assays. As virtually all Ph-like kinase fusions are associated with kinase- activated gene expression profiles, many cooperative groups and clinical laboratories have now omitted LDA screening and instead prioritized modern comprehensive testing platforms (described below) that can directly identify specific kinase fusions, which remains the essential goal of Ph-like ALL diagnostics. P2RY8-CRLF2 fusions are also known to occur in a small percentage of non–Ph-like B-ALL,20 as well as NUP214-ABL1 fusions in T-lineage acute lymphoblastic leukemia (T-ALL)21 and FIP1L1- PDGFRA fusions in myeloid neoplasms with eosinophilia,22 highlighting the potential broader applicability of such testing.
Clinical diagnostics for Ph-like ALL
| Category . | Laboratory test . | Clinical utility . | Advantages . | Disadvantages . |
|---|---|---|---|---|
| Flow cytometry | ||||
| Surface TSLPR immunophenotyping | Identification of increased TSLPR protein expression that usually correlates with CRLF2 rearrangements | Widely available testing platform Rapid turnaround time Cost-effective | Only specific for CRLF2-overexpressing B-ALL Does not distinguish between CRLF2-overexpressing Ph-like ALL vs non–Ph-like ALL | |
| FISH | ||||
| ABL1, ABL2, CSF1R/PDGFRB, PDGFRA, CRLF2, EPOR, JAK2, NTRK3 break-apart probes | Identification of known 3′ kinase genetic rearrangements in Ph-like ALL | Widely available testing platform Rapid turnaround time Cost-effective | Requires further molecular confirmation to identify specific 5′ fusion partner genes | |
| Gene expression profiling | ||||
| LDA | Detection of the kinase-activated Ph-like gene signature | Rapid turnaround time Cost-effective Capable of direct detection of P2RY8-CRLF2 fusions | Requires downstream molecular testing to identify specific kinase fusions Testing available through few reference laboratories | |
| Next-generation sequencing | ||||
| Anchored multiplex PCR-based RNA sequencing | Detection of specific gene fusions that can guide potential targeted therapy approaches | Single assay capable of identifying canonical Ph-like ALL kinase fusions Often can identify novel/previously unknown 5′ fusion partners Moderately cost-effective given comprehensive analysis (may be combined with DNA-based NGS in a single assay) | Slower time to results May miss detection of fusions involving immunoglobulin or T-cell receptor genes May not detect point mutations or insertions/deletions outside of customized gene panels | |
| Comparative genomic hybridization microarray | Genome-wide identification of copy number changes in genes relevant to hematologic malignancies | Can detect Ph-like gene fusions resulting from interstitial deletions (eg, P2RY8-CRLF2, EBF1-PDGFRB) Can detect IKZF1 and other transcription factor deletions enriched in Ph-like ALL Rapid turnaround time | Cannot detect gene fusions resulting from chromosomal translocations Relatively higher cost given limited scope of results | |
| DNA amplicon-based NGS panels | Identification of single-nucleotide variants, indels, internal tandem duplications, and copy number variations recurrently mutated in hematologic malignancies | Rapid turnaround time Can detect IKZF1 and other transcription factor deletions enriched in Ph-like ALL Can detect sequence mutations in JAK-STAT and RAS pathway genes occurring in Ph-like ALL | Does not detect gene fusions Expensive | |
| Whole transcriptome sequencing (RNA sequencing) | Comprehensive genome-wide and unbiased RNA-based analysis of gene expression, fusion transcripts, and mutations | Single assay to identify the specific gene signature, fusions and mutations of Ph-like ALL Can identify novel/previously unknown kinase alterations | Limited clinical testing availability, mostly performed in a research setting at the present time Very expensive Long time to results | |
| Category . | Laboratory test . | Clinical utility . | Advantages . | Disadvantages . |
|---|---|---|---|---|
| Flow cytometry | ||||
| Surface TSLPR immunophenotyping | Identification of increased TSLPR protein expression that usually correlates with CRLF2 rearrangements | Widely available testing platform Rapid turnaround time Cost-effective | Only specific for CRLF2-overexpressing B-ALL Does not distinguish between CRLF2-overexpressing Ph-like ALL vs non–Ph-like ALL | |
| FISH | ||||
| ABL1, ABL2, CSF1R/PDGFRB, PDGFRA, CRLF2, EPOR, JAK2, NTRK3 break-apart probes | Identification of known 3′ kinase genetic rearrangements in Ph-like ALL | Widely available testing platform Rapid turnaround time Cost-effective | Requires further molecular confirmation to identify specific 5′ fusion partner genes | |
| Gene expression profiling | ||||
| LDA | Detection of the kinase-activated Ph-like gene signature | Rapid turnaround time Cost-effective Capable of direct detection of P2RY8-CRLF2 fusions | Requires downstream molecular testing to identify specific kinase fusions Testing available through few reference laboratories | |
| Next-generation sequencing | ||||
| Anchored multiplex PCR-based RNA sequencing | Detection of specific gene fusions that can guide potential targeted therapy approaches | Single assay capable of identifying canonical Ph-like ALL kinase fusions Often can identify novel/previously unknown 5′ fusion partners Moderately cost-effective given comprehensive analysis (may be combined with DNA-based NGS in a single assay) | Slower time to results May miss detection of fusions involving immunoglobulin or T-cell receptor genes May not detect point mutations or insertions/deletions outside of customized gene panels | |
| Comparative genomic hybridization microarray | Genome-wide identification of copy number changes in genes relevant to hematologic malignancies | Can detect Ph-like gene fusions resulting from interstitial deletions (eg, P2RY8-CRLF2, EBF1-PDGFRB) Can detect IKZF1 and other transcription factor deletions enriched in Ph-like ALL Rapid turnaround time | Cannot detect gene fusions resulting from chromosomal translocations Relatively higher cost given limited scope of results | |
| DNA amplicon-based NGS panels | Identification of single-nucleotide variants, indels, internal tandem duplications, and copy number variations recurrently mutated in hematologic malignancies | Rapid turnaround time Can detect IKZF1 and other transcription factor deletions enriched in Ph-like ALL Can detect sequence mutations in JAK-STAT and RAS pathway genes occurring in Ph-like ALL | Does not detect gene fusions Expensive | |
| Whole transcriptome sequencing (RNA sequencing) | Comprehensive genome-wide and unbiased RNA-based analysis of gene expression, fusion transcripts, and mutations | Single assay to identify the specific gene signature, fusions and mutations of Ph-like ALL Can identify novel/previously unknown kinase alterations | Limited clinical testing availability, mostly performed in a research setting at the present time Very expensive Long time to results | |
TSLPR, thymic stromal lymphopoietin receptor.
Most, if not all, 3′ genes involved in canonical Ph-like ALL-associated kinase fusions can now be rapidly identified by FISH. Commercially available dual-color break-apart FISH probes for ABL-class genes ABL1, ABL2, PDGFRA, and PDGFRB (which also detects CSF1R given its adjacent location to PDGFRB) and JAK pathway genes CRLF2, JAK2, and EPOR can identify disruptions within 48 to 72 hours of ALL diagnosis. While FISH is generally not capable of identifying the 5′ fusion partners that require more detailed RNA-based molecular testing, the rapid turnaround time and wide accessibility of FISH in clinical laboratories is a highly attractive strategy to enable efficient addition of relevant TKIs during induction therapy. In addition, detection of increased thymic stromal lymphopoietin receptor cell surface expression by routine diagnostic flow cytometry immunophenotyping is universally associated with underlying CRLF2 rearrangements. This modality is another rapid (within 24-48 hours) and cost-effective method for initial identification of CRLF2-rearranged cases that comprise approximately half of Ph-like ALLs.19 Characterization of specific CRLF2 fusions or rearrangements again requires more detailed FISH or RNA-based molecular testing.
RNA-based NGS fusion assays are now frequently used in the clinic for the detection of somatic gene rearrangements in human cancers and have largely replaced prior multiplexed reverse-transcriptase polymerase chain reaction (RT-PCR) fusion panels that were initially developed for Ph-like ALL detection. As one example, the ArcherDx FusionPlex Heme panel uses anchored multiplex PCR-based enrichment to detect fusion transcripts with the ability to identify known and novel partner genes of 148 fusions occurring in lymphoid and myeloid malignancies.23 Of relevance, fusions involving immunoglobulin loci or T-cell receptor genes may not be detected by this technology, as these rearrangements often do not result in chimeric transcripts. As such, IGH-CRLF2 and IGH-EPOR rearrangements that are common in Ph-like ALL are frequently not detectable by this modality and require identification by FISH. Using similar Archer-based anchored multiplex PCR technology, the Children's Hospital of Philadelphia (CHOP) fusion panel was developed and validated to interrogate 586 different fusion transcripts amongst 106 genes in pediatric cancers with excellent sensitivity as low as 1% and is now used for routine clinical testing of all patients with newly-diagnosed or relapsed hematologic malignancies in combination with paired DNA-based tumor/normal germline NGS analysis.24 The CHOP fusion panel was recently refined to add several probes covering partner genes commonly involved in immunoglobulin gene rearrangements, including MYC, CRLF2 and EPOR, capable of additional expression elucidation and prediction of probable fusions.24 Clinical whole-transcriptome or RNA-sequencing (seq) is further becoming increasingly available and represents a powerful tool for comprehensive gene expression assessment, fusion detection, and mutation analysis.25 At present, RNA-seq is the only single platform capable of simultaneous detection of both the Ph-like expression signature by hierarchical clustering and the specific kinase fusion itself.26 Furthermore, RNA-seq provides additional information about co-occurring somatic mutations and aberrant isoforms of transcription factor genes, such as IKZF1, that are frequently altered in Ph-like ALL.26,27 Finally, the commercial FoundationOne Heme Panel offers a targeted combined DNA and RNA sequencing of >400 cancer-related genes; this combined approach may address the limitations of RNA-only-based assays in its assessment of copy number variations and single nucleotide variants.28 The above NGS assays may not be time- or cost-effective for routine analysis of all patients with ALL, but may be reserved for selected cases with unidentified kinase fusions despite high clinical suspicion. The average turn-around time for results reporting of these NGS assays ranges between two and four weeks. Another multiplex fusion assay method that may be suitable for clinical laboratories that do not have the necessary infrastructure for NGS platforms, but is less labor-cumbersome than the multiplex RT-PCR panels, is the NanoString technology. This methodology can presently detect up to 51 B-ALL fusion transcripts, requires less than one hour of hands-on time, and can deliver results within 48 hours.29
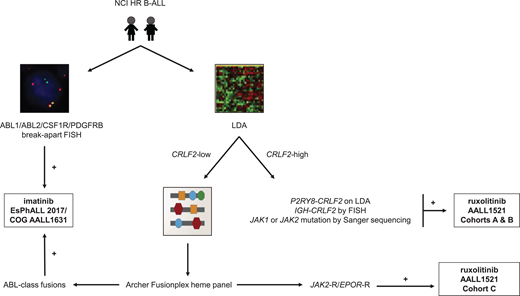
In summary, multi-modal testing is required to identify the full spectrum of genetically-heterogeneous Ph-like ALL and to allocate patients to appropriate TKI-based therapies. The advent of more comprehensive and cost-effective assays in recent years has improved diagnostic capabilities and efficiencies. Cooperative oncology groups have now established different clinical testing algorithms that have been adjusted based on available infrastructure and resources, patient volumes, and goals of care. These pragmatic Ph-like ALL diagnostic algorithms are summarized in Figure 1.
Ph-like ALL diagnostic algorithm used by the Children's Oncology Group. RT-PCR, reverse transcriptase polymerase chain reaction; TSLPR, thymic stromal lymphopoietin receptor (encoded by CRLF2).
Ph-like ALL diagnostic algorithm used by the Children's Oncology Group. RT-PCR, reverse transcriptase polymerase chain reaction; TSLPR, thymic stromal lymphopoietin receptor (encoded by CRLF2).
CLINICAL CASE 2
A 16-year-old male of Hispanic/Latinx ethnicity was diagnosed with NCI HR B-ALL after presentation with a white blood cell count of 120 000/µL, and 84% peripheral blasts and no evidence of leukemia involvement in his cerebrospinal fluid (CNS1). Diagnostic flow cytometric immunophenotyping was consistent with B-ALL and also detected increased thymic stromal lymphopoietin receptor surface staining. LDA screening showed a positive Ph-like gene signature, CRLF2 overexpression, and absence of the P2RY8-CRLF2 fusion. Cytogenetic analysis demonstrated a normal karyotype with 46,XY, and FISH testing detected an IGH-CRLF2 rearrangement. DNA-based NGS analysis further revealed deletion of IKZF1 and PAX5 and a JAK2 R683G point mutation. The patient received 4-drug induction therapy on the AALL1732 trial and had positive end-induction MRD at 0.3%. He was subsequently enrolled on the COG AALL1521 phase 2 clinical trial (NCT02723994) for postinduction chemotherapy with addition of ruxolitinib at 50 mg/m2 twice daily for 14 days on/14 days off per 28-day cycle. End-consolidation MRD was 0%. The patient continued postconsolidation chemotherapy with ruxolitinib and had anaphylaxis to peg-asparaginase in delayed intensification requiring alternative therapy with recombinant Erwinia chrysanthemi asparaginase. He remains in clinical remission and now has maintenance therapy.
The role of TKIs in Ph-like ALL
Clinical outcomes of patients with NCI HR Ph-like ALL treated with chemotherapy are universally suboptimal. Several studies have reported poor event-free survival and overall survival of patients with CRLF2-overexpressing HR B-ALL, irrespective of their Ph-like gene expression profiles, in the pre-TKI era that are comparable to those of patients with Ph-like ALL harboring ABL-class fusions.20,30,31 In contrast, outcomes of children with CRLF2-overexpressing ALL are much more favorable, although still inferior to those of children with CRLF2 wild-type standard-risk B-ALL.20,32,33 The poor outcomes of patients with HR Ph-like ALL across the age spectrum with best-available conventional chemotherapy regimens and lack of defined standard-of-care treatment approaches merit investigation of novel therapies.1,20,30
The discovery of targetable kinase-activating alterations in Ph-like ALL followed by demonstration of robust antileukemia activity of JAK pathway- or ABL-class-targeted kinase inhibitors in preclinical models provided a compelling rationale for prospective assessment of potential efficacy of TKI addition to chemotherapy in Ph-like ALL,1,4,14,15,34-37 which mirrors the highly successful treatment paradigm established for patients with Ph+ ALL.38,39 In the prior era of conventional chemotherapy without TKI intercalation, patients with Ph+ ALL had extremely high rates of treatment failure that precluded successful HSCT in CR1,30,40 which was previously the predominant curative modality for these high-risk patients. Clinical trial data subsequently demonstrated that TKI initiation ideally by mid-induction significantly reduced MRD positivity rates in pediatric patients with Ph+ ALL, which consequently reduced the indication for CR1 HSCT from 15% to 5%.38,39 Although anecdotal case reports and multipatient series have described favorable short-term outcomes of patients with Ph-like ALL who have TKI addition to chemotherapy,41-45 the potential long-term efficacy of such approaches warrants formal prospective evaluation and is under active investigation in clinical trials (Table 2).
Clinical trials for patients with Ph-like ALL
| Study group . | Trial name/phase . | Years . | Age group, y . | Genetic subgroups . | TKI . | Status . |
|---|---|---|---|---|---|---|
| COG | AALL1131/phase 3 (NCT01406756) | 2016-2019 | >1-30.99 | De novo ABL-class Ph-like B-ALL | dasatinib | Closed to accrual—pending results |
| COG | AALL1521/phase 2 (NCT02723994) | 2016-present | ≥1-21 | De novo JAK-STAT pathway-altered NCI high-risk Ph-like ALL | ruxolitinib | Cohort A: CRLF2-R and JAK+ with EOI MRD ≥0.01%—open to accrual Cohort B: CRLF2-R and JAK– with EOI MRD ≥0.01%—open to accrual Cohort C: JAK2-R, EPOR-R, and SH2B3 mutations and IL7R mutations with EOI MRD ≥0.01%—closed to accrual Cohort D: patients eligible for cohort A, B, or C with EOI MRD <0.01%—closed to accrual |
| EsPhALL + COG | EsPhALL2017/AALL1631/phase 3 (NCT03007147) | 2017-present | >1-21 | De novo Ph+ or ABL-class Ph-like ALL | Imatinib | Active |
| COG | AALL1922/phase 1/2 (NCT04501614) | 2020-present | ≥1-21 | Relapsed/refractory Ph+ or ABL-class Ph-like ALL | Ponatinib | Active |
| COG | ADVL1823/phase 2 (NCT03834961) | 2019-present | 0-≤30 | Relapsed/refractory acute leukemia with NTRK1/2/3 fusions | Larotrectinib | Cohort C—active |
| DFCI | DFCI ALL 16-001/phase 3 (NCT03040030) | 2017-present | ≥1-21 | De novo VHR B-ALL with ABL-class fusions other than BCR-ABL1 | Dasatinib | Active |
| SJCRH | Total Therapy XVII/phase 2/3 (NCT03117751) | 2017-present | ≥1-18 | De novo B-ALL with ABL-class fusions or JAK-STAT pathway mutations | Dasatinib, ruxolitinib | Active |
| ALLTogether | ALLTogether 01/phase 3 (NCT03911128) | 2019-present | ≥1-45 | De novo Ph-like ABL-class B-ALL | Imatinib | Active |
| MDACC | 2014-0521/phase 1/2 (NCT02420717) | 2015-2021 | ≥10 | relapsed/refractory Ph-like ALL with ABL-class or JAK-STAT pathway alterations | Dasatinib, ruxolitinib | Closed to accrual |
| University of Chicago | IRB17-1110/phase 1 (NCT03571321) | 2019-present | ≥18-39.99 | De novo Ph-like ALL harboring JAK-STAT pathway alterations | Ruxolitinib | Active |
| SWOG | S1318/phase 2 (NCT02143414) | 2015-2017 | ≥65 | De novo B-ALL with ABL-class fusions including BCR-ABL1 | Dasatinib | Cohort 2—closed to accrual |
| Study group . | Trial name/phase . | Years . | Age group, y . | Genetic subgroups . | TKI . | Status . |
|---|---|---|---|---|---|---|
| COG | AALL1131/phase 3 (NCT01406756) | 2016-2019 | >1-30.99 | De novo ABL-class Ph-like B-ALL | dasatinib | Closed to accrual—pending results |
| COG | AALL1521/phase 2 (NCT02723994) | 2016-present | ≥1-21 | De novo JAK-STAT pathway-altered NCI high-risk Ph-like ALL | ruxolitinib | Cohort A: CRLF2-R and JAK+ with EOI MRD ≥0.01%—open to accrual Cohort B: CRLF2-R and JAK– with EOI MRD ≥0.01%—open to accrual Cohort C: JAK2-R, EPOR-R, and SH2B3 mutations and IL7R mutations with EOI MRD ≥0.01%—closed to accrual Cohort D: patients eligible for cohort A, B, or C with EOI MRD <0.01%—closed to accrual |
| EsPhALL + COG | EsPhALL2017/AALL1631/phase 3 (NCT03007147) | 2017-present | >1-21 | De novo Ph+ or ABL-class Ph-like ALL | Imatinib | Active |
| COG | AALL1922/phase 1/2 (NCT04501614) | 2020-present | ≥1-21 | Relapsed/refractory Ph+ or ABL-class Ph-like ALL | Ponatinib | Active |
| COG | ADVL1823/phase 2 (NCT03834961) | 2019-present | 0-≤30 | Relapsed/refractory acute leukemia with NTRK1/2/3 fusions | Larotrectinib | Cohort C—active |
| DFCI | DFCI ALL 16-001/phase 3 (NCT03040030) | 2017-present | ≥1-21 | De novo VHR B-ALL with ABL-class fusions other than BCR-ABL1 | Dasatinib | Active |
| SJCRH | Total Therapy XVII/phase 2/3 (NCT03117751) | 2017-present | ≥1-18 | De novo B-ALL with ABL-class fusions or JAK-STAT pathway mutations | Dasatinib, ruxolitinib | Active |
| ALLTogether | ALLTogether 01/phase 3 (NCT03911128) | 2019-present | ≥1-45 | De novo Ph-like ABL-class B-ALL | Imatinib | Active |
| MDACC | 2014-0521/phase 1/2 (NCT02420717) | 2015-2021 | ≥10 | relapsed/refractory Ph-like ALL with ABL-class or JAK-STAT pathway alterations | Dasatinib, ruxolitinib | Closed to accrual |
| University of Chicago | IRB17-1110/phase 1 (NCT03571321) | 2019-present | ≥18-39.99 | De novo Ph-like ALL harboring JAK-STAT pathway alterations | Ruxolitinib | Active |
| SWOG | S1318/phase 2 (NCT02143414) | 2015-2017 | ≥65 | De novo B-ALL with ABL-class fusions including BCR-ABL1 | Dasatinib | Cohort 2—closed to accrual |
CRLF2-R, CRLF2-rearranged; DFCI, Dana-Farber Cancer Institute; EOI, end of induction; EPOR-R, EPOR-rearranged; JAK+, JAK2- or JAK1-mutated; JAK–, JAK2 and JAK1 wild-type; JAK2-R, JAK2-rearranged; MDACC, MD Anderson Cancer Center; SJCRH, St Jude Children's Research Hospital; SWOG, Southwest Oncology Group; VHR, very high risk.
The JAK1/2 inhibitor ruxolitinib is currently being studied in clinical trials for patients with Ph-like ALL harboring CRLF2 rearrangements or other JAK pathway alterations. In the COG ADVL1011 phase 1 trial, the safety and tolerability of ruxolitinib monotherapy was demonstrated in children with multiply-relapsed/refractory cancers, and a recommended phase 2 dose (RP2D) of 50 mg/m2 twice daily for 28 days/cycle was identified.46 In the subsequent COG AALL1521 phase 2 trial, a part 1 safety phase evaluated addition of 6 different dose levels of ruxolitinib to multiagent chemotherapy in children and adolescents and young adults with newly diagnosed Ph-like ALL and identified an RP2D of ruxolitinib 50 mg/m2 twice daily given intermittently on a 14 days on/14 days off schedule in combination with chemotherapy.47 Increased frequency or severity of adverse events with ruxolitinib addition was not observed over rates seen in patients treated with backbone chemotherapy.47 Although the results of AALL1521 are not yet available from the subsequent part 2 efficacy phase of patients treated uniformly at the RP2D, favorable outcomes have been reported to date in case reports of patients with Ph-like ALL harboring CRLF2, JAK2, or EPOR rearrangements treated with ruxolitinib and chemotherapy.44,45,48
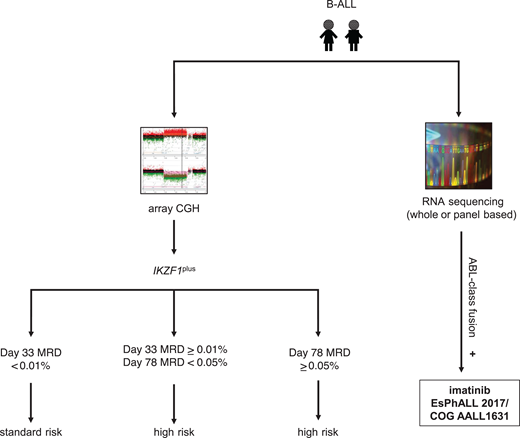
At present, 3 kinase inhibitors have been or are under current clinical evaluation in pediatric patients with ABL-class Ph-like ALL. The international EsPhALL 2017/COG AALL1631 phase 3 trial for patients with Ph+ ALL was recently amended to extend eligibility to patients with Ph-like ALL harboring an ABL-class fusion. This randomized study, aimed at toxicity reduction while preserving antileukemia efficacy, is testing imatinib with more intensive standard-of-care EsPhALL therapy (control arm A) vs less intensive COG HR B-ALL (experimental arm B) chemotherapy backbones.49 The AIEOP-BFM ALL 2017 trial (NCT03643276) previously allowed patients with identified ABL-class Ph-like ALL to be treated with imatinib and the control chemotherapy arm of their respective treatment assignment prior to amendment of the EsPhALL 2017/COG AALL1631 trial (Figure 2). The ALLTogether-01 trial (NCT03911128) also includes patients with ABL-class Ph-like fusions in the overall study for imatinib addition to chemotherapy and risk stratification according to protocol-predefined criteria. Previously, the second-generation SRC/ABL inhibitor dasatinib with chemotherapy was investigated in a descriptive cohort of pediatric patients with ABL-class Ph-like ALL (n = 28) via the COG AALL1131 phase 3 trial (NCT02883049), but outcomes data have not yet been released. Finally, the third-generation ABL-class inhibitor ponatinib, which is effective against the ABL1 T315I resistance mutation, is currently under study with chemotherapy for children with relapsed/refractory Ph+ ALL or ABL-class Ph-like ALL via the COG AALL1922 phase 2 trial (NCT03934372).
Ph-like ALL diagnostic algorithm used by the AIEOP-BFM consortium. CGH, comparative genomic hybridization; IKZF1plus, deletion of IKZF1 and other B-cell transcription factors as defined by Stanulla et al.56
Ph-like ALL diagnostic algorithm used by the AIEOP-BFM consortium. CGH, comparative genomic hybridization; IKZF1plus, deletion of IKZF1 and other B-cell transcription factors as defined by Stanulla et al.56
Antibody-based and cellular immunotherapy has transformed the therapeutic landscape of relapsed/refractory B-ALL and is now being investigated in patients with newly diagnosed ALL via frontline trials. Blinatumomab or inotuzumab monotherapy has shown appreciable activity in adults with relapsed/refractory Ph-like ALL among other leukemia subtypes.50,51 Recently, the combination of immunotherapy with TKI to deepen response and/or reduce treatment-related toxicities from traditional intensive chemotherapy has also been explored in clinical trials. Early results of the D-ALBA phase 2 trial for adults with newly diagnosed Ph+ ALL evaluating a chemotherapy-free regimen, consisting of 3-month induction with dasatinib and prednisone followed by consolidation with ≥2 cycles of blinatumomab and dasatinib, yielded outstanding 1-year disease-free and overall survival of 88% and 95%, respectively.52 Blinatumomab with dasatinib53 or ponatinib54 and inotuzumab with bosutinib55 represent additional combination immunotherapy/TKI approaches with early promise in adults with ABL-class Ph-like and/or Ph+ ALL that may warrant further clinical investigation also in children and adolescents/young adults. To this end, the randomized EsPhALL 2022/COG AALL2131 phase 3 trial of imatinib with chemotherapy vs chemotherapy and blinatumomab is in development for patients with ALL harboring BCR-ABL1 or other ABL-class rearrangements (T. H. Tran and S. K. Tasian, personal communication, 2022). In conclusion, the historically poor outcomes of patients with Ph-like ALL may be overcome by early TKI introduction to deepen initial MRD-negative remission rates and reduce indication for HSCT in CR1, followed by protracted TKI administration with different chemoimmunotherapy cassettes under current clinical evaluation with an overarching goal of reducing relapse risk and improving long-term cure.
Acknowledgments
These studies were supported by the National Institutes of Health (NIH)/National Cancer Institute awards 1U01CA232486 and U01CA243072 (SKT), Department of Defense Translational Team Science award CA180683P1 (SKT), V Foundation for Cancer Research (SKT), and Leukemia and Lymphoma Society (Scholar Award to SKT). SKT is the Joshua Kahan Endowed Chair in Pediatric Leukemia Research at the Children's Hospital of Philadelphia.
Conflict-of-interest disclosure
Thai Hoa Tran received research funding from Servier and serves or has served on the scientific advisory boards of Bayer Pharmaceutics, Jazz Pharmaceuticals, and Servier Pharmaceuticals.
Sarah K. Tasian receives research funding from Beam Therapeutics, Incyte Corporation (for Ph-like ALL studies), and Kura Oncology and serves or has served on the scientific advisory boards of Aleta Biotherapeutics, bluebird bio, GSK, Kura Oncology, and Syndax Pharmaceuticals.
Off-label drug use
Thai Hoa Tran: off-label use of tyrosine kinase inhibitors (dasatinib, imatinib, ruxolitinib) for patients with Ph-like ALL is discussed.
Sarah K. Tasian: off-label use of tyrosine kinase inhibitors (dasatinib, imatinib, ruxolitinib) for patients with Ph-like ALL is discussed.