Abstract
In recent years, the composite molecular architecture in acute myeloid leukemia (AML) has been mapped out. We now have a clearer understanding of the key genetic determinants, the major genetic interactions, and the broad order in which these mutations occur. The next impending challenge is to discern how these recent genomic discoveries define disease biology as well as how to use molecular markers to deliver patient-tailored clinical decision support.
Learning Objectives
To obtain a broad understanding of the molecular pathogenesis of acute myeloid leukemia
To relate how the underlying molecular structure segregates distinct biological and subgroups in acute myeloid leukemia and how these factors in turn associate with distinct clinical features such as age at presentation and outcomes
To assess the main challenges and future opportunities for the incorporation of molecular markers in clinical algorithms for acute myeloid leukemia
Introduction
The identification of recurrent cytogenetic abnormalities associated with distinct clinical presentation in acute myeloid leukemia (AML) paved the way for the incorporation of genetic markers into clinical decision-making.1,2 The observation that most cytogenetic abnormalities are nonoverlapping and their distinct associations with clinical presentation (age, white blood cell counts, and morphology), therapeutic response (attainment of complete remission after induction), relapse rates, and overall survival formed the basis for the development of molecular classification and risk stratification schemas by the World Health Organization (WHO) and the European Leukemia Net (ELN).2-5
These schemas have significantly improved clinical management in leukemia.2-5 For example, understanding the molecular basis of chronic myelogenous leukemia and acute promyelocytic leukemia has fundamentally changed the clinical outlook for these 2 disease subsets. The identification of BCR-ABL and PML-RARA fusion genes as the 2 causative lesions for chronic myelogenous leukemia and acute promyelocytic leukemia, respectively, delivered a robust genetic biomarker for diagnosis and led to the development of highly efficacious and well-tolerated therapeutic interventions, turning 2 lethal diseases into subsets that show remarkable long-term patient survival.6-8 These paradigms highlight how defining the molecular basis of disease pathogenesis can identify new disease subsets, alter clinical management, and deliver curative treatments. Taken together, these have set the bar high for further pursuits of patient-tailored medicine in leukemia.
With an ever-increasing understanding of the molecular pathogenesis of leukemia, the WHO and ELN convene regularly to review histopathologic and genomic findings2-5 and to update recommendations on diagnosis, prognosis, and associated clinical management in accordance with current knowledge. The aim is to generate internationally adaptable, practical, and accessible prognostic categories on the basis of substantial and independently validated evidence of clinical and prognostic associations.
Current challenges in AML
Beyond cytogenetic markers, disease classification and, importantly, risk stratification remain challenging for ∼50% of patients with AML who present with normal karyotypes.1,2 Conventionally, normal karyotype AML (NK-AML) has been associated with favorable or intermediate-risk disease. However, although most patients respond to induction chemotherapy, relapse is frequent, and clinical response within this subgroup has been extremely variable, making this group of patients one of the most challenging to risk-stratify and treat. A second group of patients that is challenging to stratify and derive treatment decisions for is represented by older patients (ie, those aged >60 years).
Age is one of the strongest contributing risk factors for the development of AML, with ∼74% of patients with AML presenting at an age ≥55 years (US Surveillance, Epidemiology, and End Results data; https://seer.cancer.gov/). In multivariate analysis, age represents one of the most adverse prognostic indicators for response to treatment and overall survival.9 Although changes in treatment scheduling and stem cell transplantation have shown some benefit in younger patients with AML, survival in older individuals remains dismal. Decisions on dose intensification and transplant rely entirely on appropriate risk stratification.
Genome sequencing in AML
Sequencing of AML genomes found that AML is genetically diverse and clonally heterogeneous, with multiple mutations acquired over time, and complex patterns of clonal evolution shaping disease progression and response to therapy.10-12 As the mystique of the AML genome is unraveled, the complex patterns of mutation acquisition seen in NK-AML reveal a more heterogeneous genomic landscape that cannot be readily substratified into nonoverlapping molecular and clinical entities.10 Even for the well-versed AML community, which has been at the forefront of developing molecular-guided classification schemas, the complexity of AML genomes, the observation that most patients harbor multiple gene mutations, and the dynamic patterns of disease evolution impose significant challenges and considerations that need to be addressed.10,11,13-15 The present review discusses recent molecular discoveries in AML, how these help us understand AML pathogenesis, and important considerations for the inclusion of such findings into future classification and prognostication schemas.
Mutation acquisition in AML
The incidence of AML increases with age, which ties in well with our current understanding of mutation acquisition in time. Sequencing studies of normal hematopoietic stem and progenitor clones derived from healthy individuals show a stable increase of acquired mutations, consistent with acquisition of one coding mutation per decade of life.16 Reflecting this observation, population studies of peripheral blood cells from healthy individuals show an age-dependent increase of acquired mutations in the blood.17-20 Thus, with age, the incidence of stably acquired mutations in the bone marrow increases, as does the probability of acquiring a mutation in a gene that will confer selective advantage and clonal selection in hematopoietic stem and progenitor cells.
The AML genome
Analysis of 200 de novo AML patients according to whole-genome or whole-exome sequencing identified an excess of 200 recurrently mutated genes, of which 23 were significantly mutated.13 Although patients with AML often share mutations seen in normal healthy individuals with clonal hematopoiesis17-20 (representing, on average, <10% of cells in the blood), in AML, most patients have ≥2 acquired mutations and are clonally represented (∼100% of cells).
Among the gene mutations described, AML has both a common and a distinct repertoire of driver gene mutations, many of which fall into shared functional classes. Of its own panoply of specific oncogenic drivers, mutations that lead to aberrant regulation of DNA methylation and hydroxymethylation (DNMT3A, TET2, and IDH1/2), altered messenger RNA splicing by the U2 complex (SF3B1, SRSF2, and U2AF1) modified chromatin architecture (ASXL1, EZH2, and KMT2A), and transcriptional deregulation (CEBPA, RUNX1, and WT1) are most frequent.21 The nuclear-shuttling factor NPM1 is mutated with high frequency (∼30%), and mutations in components of the cohesin complex (SMC1, SMC3, STAG2, and RAD21) have been described for the first time in AML.13
In common with nearly all cancer indications, AML is frequently driven by acquisition of ligand-independent proliferative and prosurvival signal transduction. This action occurs either at the level of transmembrane receptors such as FLT3 or KIT, deactivation of inhibitory phosphatases (PTPN11), and also by activating mutations in constitutive transduction factors such as NRAS and (less frequently) KRAS. These events are typically acquired late in leukemic development. In tandem with strong mitogenic signaling, cell cycle inhibitory tumor suppressors such as TP53 are also frequently lost.
Patterns of genomic instability allude to distinct evolutionary trajectories and define molecular and clinical subtypes in AML
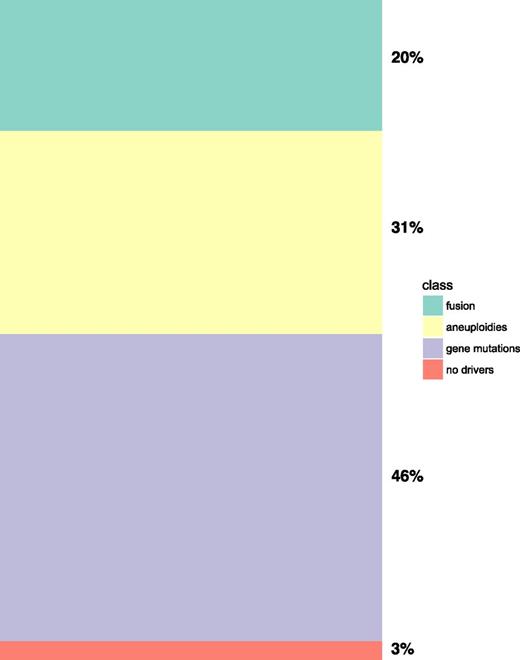
This first analysis of AML genomes showed 3 broad patterns of genomic instability: (1) AML with balanced genomic rearrangements or fusion genes; (2) AML with chromosomal aneuploidies; and (3) AML with normal karyotypes dominated by gene mutations (Figure 1). More recent genome profiling analysis across the AML subtypes revealed that these broad patterns of genomic instability are accompanied by very specific and ordered patterns of co-mutations. The specificity of these interactions, however, suggests a strong dependency for co-mutation indicative of functional co-operativity at the cellular level during AML development13,23 (Figure 2A-C).
Meta-analysis of Papaemmanuil et al23 2016 data for broad patterns of genomic instability in AML shows that ∼20% of patients with AML were defined according to fusion genes, 31% by chromosomal aneuploidies (to include at least 1 aneuploidy), 46% by gene mutations only (in the absence of gene fusions and chromosomal aneuploidies), and 3% with no events.
Meta-analysis of Papaemmanuil et al23 2016 data for broad patterns of genomic instability in AML shows that ∼20% of patients with AML were defined according to fusion genes, 31% by chromosomal aneuploidies (to include at least 1 aneuploidy), 46% by gene mutations only (in the absence of gene fusions and chromosomal aneuploidies), and 3% with no events.
Distribution of number of acquired mutations according to AML class and most frequently co-mutated genes. Data shown for (A) AML with fusion genes, (B) AML with aneuploidies, and (C) AML with gene mutations.
Distribution of number of acquired mutations according to AML class and most frequently co-mutated genes. Data shown for (A) AML with fusion genes, (B) AML with aneuploidies, and (C) AML with gene mutations.
AML with balanced genomic rearrangements
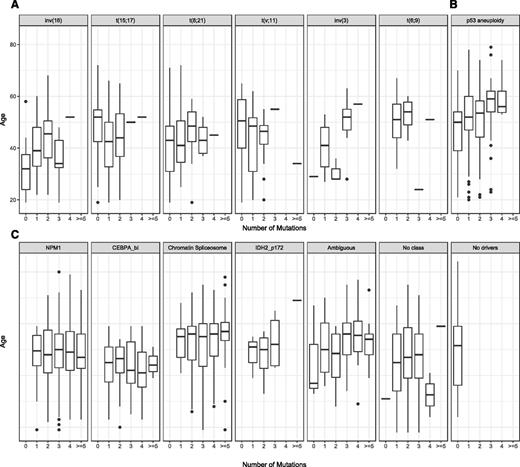
AML with balanced genomic rearrangements or fusion genes had, on average, 1 genomic rearrangement and a low number of other gene mutations, most frequently implicating activating mutations in signaling genes to include NRAS, FLT3, KIT, tyrosine, or serine– threonine kinases and protein tyrosine phosphatases.13 Strikingly, most patients with AML with fusion genes tend to present at a younger age and have relatively simple genomes, with lower overall number of acquired mutations (Figure 3A). There are at least 8 distinct molecular subtypes that are defined by recurrent genomic rearrangements and are recognized by the WHO as distinct clinicopathologic entities. Each of these affect 1% to 10% of AML patients, respectively, and include the following: PML-RARA, defined by t(15;17)(q22;q21); RUNX1-RUNX1T1, defined by t(8;21)(q22;q22.1); CBFB-MYH11, defined by inv(16)(p13.1q22) or t(16;16)(p13.1;q22); MLLT3-KMT2A, defined by t(9;11)(p21.3;q23.3); DEK-NUP214, defined by t(6;9)(p23;q34.1); GATA2 MECOM, defined by inv(3)(q21.3q26.2)/t(3;3)(q21.3;q26.2); and RBM15-MKL1, defined by (t1;22)(p13.3;q13.3). This topic was reviewed in Bullinger et al.24
Distribution of number of acquired mutations according to age of diagnosis for each AML class ordered as (A) AML with fusion genes, (B) AML with aneuploidies, and (C) AML with gene mutations.
Distribution of number of acquired mutations according to age of diagnosis for each AML class ordered as (A) AML with fusion genes, (B) AML with aneuploidies, and (C) AML with gene mutations.
Prognostic implications
Most AML subtypes defined according to genomic re-arrangements have distinct and relatively uniform clinical outcomes. This scenario may be, in part, accounted for by the simple underlying genomic architecture observed in these subtypes. Accounting for the frequent co-operating lesions within each fusion gene subtype may identify significant modifiers of therapeutic response and overall survival.24 This has been well-described for RUNX1-RUNX1T1 AML, which, when co-mutated with KIT mutations, turns a favorable disease to one associated with inferior prognosis.1 Although we have begun to identify which genes preferentially co-occur with one another,13,22,25 our ability to study the clinical impact of such co-occurrences remains limited.
AML with chromosomal aneuploidies
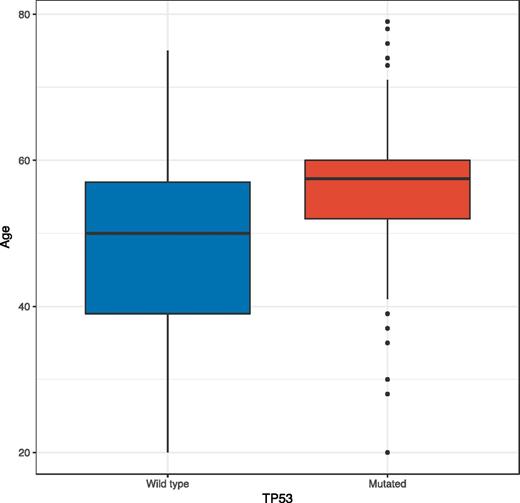
Contrary to AML subtypes that are defined according to balanced genomic rearrangements, AML with chromosomal aneuploidy represents a markedly more heterogeneous subgroup. The majority (>60%) of cases in this subgroup have complex karyotypes, defined here as ≥3 chromosomal events. Of these, the most frequent chromosomal events are −5/5q, −7/7q, −17/17p, −12/12p, and, to a lesser extent, +8/8q. Approximately 50% of patients in this subgroup have mutations in TP53 (Figure 2B). Even within this homogenously defined group, TP53 mutations are strongly correlated with old age: the median age of patients with chromosomal aneuploidy and TP53 mutations is 58 years as opposed to 49 years for the patients with aneuploidy alone (Figure 4).
Box plot indication age of diagnosis for all patients in AML with chromosomal aneuploidies and TP53 mutations, separated by TP53 mutations status, shows that average age of TP53 wild type is 49 years while TP53 mutated is 58 years.
Box plot indication age of diagnosis for all patients in AML with chromosomal aneuploidies and TP53 mutations, separated by TP53 mutations status, shows that average age of TP53 wild type is 49 years while TP53 mutated is 58 years.
Notably, the molecular and clinical presentation in this subgroup mirrors the molecular and clinical characteristics of complex karyotype myelodysplastic syndromes; it include mutations in TP53, similar chromosomal involvement, and presentation with low blast counts and older age.23 However, these 2 analogous subgroups are considered as distinct entities by the recent WHO classification of myeloid neoplasms and importantly are diagnosed and treated differently. It remains an open question whether AML with chromosomal aneuploidy is truly distinct from myelodysplastic syndromes with chromosomal aneuploidy. Studies evaluating the comprehensive spectrum of acquired mutations and how these factors relate to individual clinicopathologic features are needed. This topic is of particular interest given recent data suggesting that TP53 mutated and complex karyotype AML may respond favorably to hypomethylating agents.26,27
Prognostic implications
Overall prognosis is reduced with the higher number of chromosomal involvement (or complex karyotype). Although patients with either complex karyotype or TP53 mutations are each associated with poor overall outcomes, patients with both TP53 and complex karyotype demarcate a subgroup of patients, with significantly inferior outcomes to either subset alone.23,28 Thus, in addition to cytogenetic profiling, testing for TP53 mutations at diagnosis is becoming increasingly important.
NK-AML with gene mutations
NK-AML accounts for 50% of AML and represents the third and largest broad cytogenetic category in AML. This category segregates in distinct and nonoverlapping mutational groups: NPM1 mutated, bi-allelic CEBPA, and the chromatin-spliceosome group.
NPM1-mutated AML
Mutations in NPM1 represent a cardinal genomic alteration in normal karyotype AML and account for 30% of AML overall. Notably, mutations in NPM1 are mutually exclusive to other genomic rearrangements and/or AML with chromosomal aneuploidy. This observation, coupled with the distinct morphology at presentation and overall favorable outcomes, led to the proposition of NPM1-mutated AML as a distinct AML class in 2017. Patients in this subgroup are typically younger and exhibit a specific and ordered pattern of co-mutations. Mutations in DNA hydroxymethylation genes (DNMT3A, TET2, IDH1, and IDH2) are reported in 75% of NPM1-mutated AML and typically represent the first acquired event, found in 100% of the leukemic cells. NPM1 is acquired as a secondary event, together with mutations in FLT3, NRAS, and PTPN11 (Figure 2B). The recently described mutations in cohesin complex genes such as RAD21, SMC3, and SMC1A and the cluster of hotspot mutations in MYC also co-segregate with NPM1 mutations.23
Prognostic implications
A recent study has shown that the broad spectrum of co-mutations in this subgroup influences both clinical presentation and outcomes. Patients with mutations in RAD21 and/or NRAS codon 12/13 mutations exhibit extremely favorable outcomes,13,23 whereas patients with NPM1 mutations and IDH1 or IDH2 mutations show increased rates of refractory disease and a tendency toward higher relapse rates.23 Last, patients with concomitant mutations in NPM1, DNMT3A, and FLT3ITD, which represent the most frequent triple genotype in AML, have significantly inferior overall survival, shorter event-free survival, higher proportion of relapse disease, and relapse-related mortality.
NPM1-mutated AML represents a paradigm of how the dense co-mutation structure molds clinical presentation and shapes treatment response. Although further stratification of NPM1-mutated AML on the basis of FLT3ITD has become common practice, further revisions to this schema will be required in the near future. This approach will be particularly important for the co-occurring IDH1 and IDH2 mutations or the more recently described adverse prognostic genes ASXL1 and RUNX1, which are currently not accounted for by the ELN when they present within a favorable prognostic group.5
Bi-allelic CEBPA
The second subset of NK-AML that is recognized by the WHO and the ELN is that defined by bi-allelic CEBPA mutations. Similar to AML with balanced genomic rearrangements, bi-allelic CEBPA AML is frequent in young patients and represents a genetically and clonally simple disease. GATA2 and NRAS mutations are found in ∼30% of patients, and mutations in WT1 as well as CSF3R are also frequent (∼20%)24 (Figure 2C). The effect on clinical presentation and outcome for these mutations is less clear. Such information can highlight new therapeutic avenues that are specific to the biology and the genetic dependencies in bi-allelic CEBPA AML.29 Intriguingly, NPM1 and bi-allelic CEBPA present at a young age and do not exhibit a correlation between mutation acquisition and age (Figure 3C). Beyond NPM1 and bi-allelic CEBPA AML, most NK-AML presents later in life (>40 years of age) and is genetically and clonally more heterogeneous (Figures 2-3).
Chromatin-spliceosome AML
Two recent studies proposed RUNX1, a transcription factor mutated in ∼8% of AML, as a subgroup-defining alteration.22,30 Mutations in RUNX1 are mutually exclusive to NPM1, CEBPA-biallelic, and AML with recurrent cytogenetic abnormalities. Patients with RUNX1 mutations present typically with older age and have lower overall survival. These 3 observations rendered RUNX1 as a good candidate for a distinct molecular subgroup in AML. However, RUNX1-mutated AML is genetically very heterogeneous. RUNX1 mutations follow complex patterns of co-mutation involving other chromatin modifiers, transcription factors, and members of the spliceosome complex that are frequently mutated in RUNX1 wild-type AML.22,23,30,31 Using agnostic classifier models, we recently proposed a broader AML class that encapsulates RUNX1-mutated AML but is not exclusive to the provisional RUNX1 category; we termed this category the chromatin-spliceosome class.23
AML with chromatin and splicing factor gene mutations represent the second largest (∼20%) molecular group in AML. Within this group, there is no single defining event but rather a shared pattern of co-mutated genes frequently implicating at least 1 epigenetic modifier, transcription factor, and/or splicing factor gene. ASXL1, RUNX1, or splicing factor genes (SF3B1, SRSF2, U2AF1, and ZRSR2), STAG2 and MLL partial tandem duplication (MLLPTD) represent the most frequently mutated genes exclusive to this class (Figure 2C). With current sample sizes, it is hard to dissect independent clinical and prognostic associations for each gene within this subgroup. Nonetheless, all molecular subsets (ASXL1 mutated, RUNX1 mutated, and splicing factor mutated) share common clinical presentation, defined by older age (median age, 58 years), lower blast counts, and enrichment of, but not profound, dysplasia.
Prognostic implications
In univariate and multivariate analysis, many of the contributing genes within this subgroup (ASXL1, RUNX1, SRSF2, SF3B1, STAG1, MLLPTD, and EZH2) independently associate with poor outcomes.23 Importantly, in this class, mutation number increases with older age (Figure 3), and prognosis steadily decreases with increasing number of mutations.
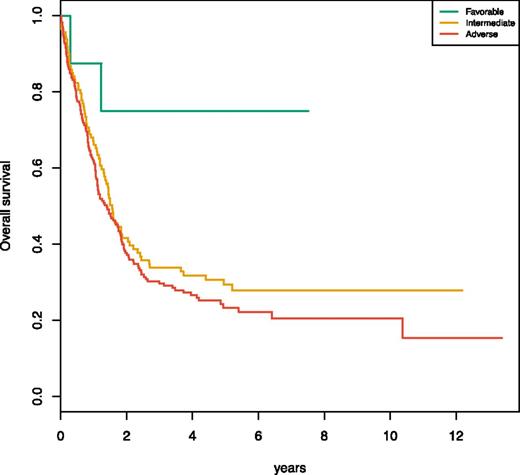
The definition of the chromatin-spliceosome group as a distinct AML class has significant clinical implications. The majority of patients in this group were stratified as intermediate risk de novo AML according to the previous ELN stratification. Data from the AML study group (AML-SG) study23 clearly show that this molecular subset demarcates a previously unrecognized adverse prognostic subset that has low complete remission rates and high relapse-related mortality. In the 2017 ELN revision, 2 major contributors to this class, ASXL1 and RUNX1, are recognized as adverse prognostic markers in the absence of a favorable prognostic lesion. However, under the 2017 criteria using the AML-SG dataset, 60% of the patients in this group would be recognized as adverse risk, 38% as intermediate risk, and 2% would overlap with favorable features. Here, we show that the patients in this class who would currently be classified as intermediate risk also have an unfavorable prognosis (Figure 5). Larger studies will further validate this molecular and clinical entity and importantly should enable incorporation of class representative genes for this group into formal stratification algorithms for AML.
Meta-analysis of the Papaemmanuil et al23 2016 chromatin-spliceosome subgroup. Kaplan-Meier curves for overall survival are shown for 3 subsets of patients that classify with the chromatin-spliceosome group (n = 299) substratified according to ELN 2017 risk (favorable, n = 8; intermediate, n = 113; and adverse, n = 178). As shown by the graphic, the 113 patients who are classified as intermediate risk have equally adverse outcomes as the patients classified as adverse risk according to RUNX1, ASXL1, or FLT3ITD high or chromosomal aneuploidies.
Meta-analysis of the Papaemmanuil et al23 2016 chromatin-spliceosome subgroup. Kaplan-Meier curves for overall survival are shown for 3 subsets of patients that classify with the chromatin-spliceosome group (n = 299) substratified according to ELN 2017 risk (favorable, n = 8; intermediate, n = 113; and adverse, n = 178). As shown by the graphic, the 113 patients who are classified as intermediate risk have equally adverse outcomes as the patients classified as adverse risk according to RUNX1, ASXL1, or FLT3ITD high or chromosomal aneuploidies.
AML with IDHR172 mutations in the absence of other class-defining genes
In the AML-SG study, a new small (1%) class was proposed, defined by using mutations in IDH2R172 in the absence of other class-defining lesions.23 This group associates with favorable prognosis and warrants validation in larger studies.
Insights into the molecular underpinnings of high-risk and older AML
Older AML, defined as that in patients >60 years of age, has been one of the most challenging subsets to treat.9 The adverse prognosis chromatin-spliceosome subgroup, AML with chromosomal aneuploidy, and TP53 mutations are enriched in older AML (49% of >60 year-olds belong to either group) and maintain independent adverse prognostic potential when age is accounted for (Figure 3C). Thus, the high risk conventionally associated with older age is in part explained by the aggregation of poor cytogenetics and, importantly, gene mutations associated with adverse outcomes. This finding has important implications on how we manage and treat older patients with AML. Derivation of risk estimates on the basis of age and the genetic profile of each patient may identify patient subsets that are aged >60 years without adverse genetics, and these patients could potentially benefit from escalated treatment regiments.
Molecular testing
Molecular testing for disease-classifying and prognostic markers as defined by the ELN is becoming routine in clinical practice. Variability in clinical outcomes across and within the prognostic groups represents one of the major clinical challenges faced by AML clinicians today. Recent data show that a large proportion of this variability is determined by the composite genomic architecture of the disease.28 Thus, it is becoming increasingly important to extend genetic testing at diagnosis to a broader array of genes implicated in AML pathogenesis and associated with clinical presentation and outcomes. Importantly, among the genes recurrently mutated in AML, we see an increasing number of approved and investigational therapeutic targets to include FLT3, IDH1, IDH2, NRAS, KRAS, PTPN11, EZH2, and mutations in the spliceosome machinery. Extending molecular testing to these putative therapeutic targets will support upfront enrollment to clinical trial protocols.
Future directions
Understanding the genomics of AML represents both an opportunity and a challenge. On one end, we now have the first blueprint for AML development, which we can use to guide the design of patient-relevant models of disease biology, as well as rationalize diagnostic, prognostic, disease surveillance, and combination therapy protocols. On the other end, it is becoming increasingly clear that using a single diagnostic and prognostic marker is not sufficient to fully inform disease state and prognosis. With increasing adoption of molecular profiling at diagnosis, this information is becoming readily available within the first weeks of a patient’s diagnosis. The next impeding challenge for the AML community lies in understanding how to interpret this information and apply it practically in the clinic. The dimensionality of the number of variables and variable combinations seems daunting. With large collaborative consortia, however, we can assemble sample sizes that are statistically powered to characterize the minimal set of gene or genotype combinations that deliver the highest prognostic value for inclusion into clinical algorithms globally.
For example, although patients with AML have, on average, 3 acquired mutations (range, 0-9), current stratification algorithms consider just 1 mutation in 89% of patients and 2 in 11%. The FLT3ITD substratification of NPM1-mutated AML accounts for all cases in which 2 mutations are considered. This finding is in stark contrast to the number of independent gene mutations we find in each patient at diagnosis. In time, we will and—importantly—should be in a position to interpret the clinical implications of the most frequent genotype combinations observed in patients with AML. Thus, using the composite molecular and clinical variables that define a patient’s leukemia, we will be in a position to derive accurate and end point–specific (attainment of complete remission, relapse, and overall survival) risk estimates for each patient.32 Informed by the genomic architecture of AML, we can design statistically powerful and population-based correlative studies and develop experimental models that recapitulate the genetic diversity and clonal heterogeneity seen in patients with AML. By considering these recent discoveries and accounting for the genetic structure of this disease, the AML community will be in a position to provide the necessary evidence for incorporation of these recent gene discoveries into advanced prognostic algorithms to support patient-tailored medicine in leukemia.
Acknowledgments
The authors thank H. Döhner and all members of the AML-SG for allowing us to reproduce figures from the data generated in Papaemmanuil et al.
Correspondence
Elli Papaemmanuil, Department of Epidemiology-Biostatistics, Center for Heme Malignancies, Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, 1275 York Ave, Box 20, New York, NY 10065; e-mail: papaemme@mskcc.org.
References
Competing Interests
Conflict-of-interest disclosure: The authors declare no competing financial interest.
Author notes
Off-label drug use: None disclosed.