Abstract
In excess of 340 blood group antigens have now been described that vary between individuals. Thus, any unit of blood that is nonautologous represents a significant dose of alloantigen. Most blood group antigens are proteins, which differ by a single amino acid between donors and recipients. Approximately 1 out of every 70 individuals are transfused each year (in the United States alone), which leads to antibody responses to red blood cell (RBC) alloantigens in some transfusion recipients. When alloantibodies are formed, in many cases, RBCs expressing the antigen in question can no longer be safely transfused. However, despite chronic transfusion, only 3% to 10% of recipients (in general) mount an alloantibody response. In some disease states, rates of alloimmunization are much higher (eg, sickle cell disease). For patients who become alloimmunized to multiple antigens, ongoing transfusion therapy becomes increasingly difficult or, in some cases, impossible. While alloantibodies are the ultimate immune effector of humoral alloimmunization, the cellular underpinnings of the immune system that lead to ultimate alloantibody production are complex, including antigen consumption, antigen processing, antigen presentation, T-cell biology, and B-cell biology. Moreover, these cellular processes differ to some extent with regard to transfused RBCs as compared with other better-studied immune barriers (eg, infectious disease, vaccines, and solid organ transplantation). The current work focuses on illustrating the current paradigm of humoral immunity, with a specific focus on particulars of RBC alloimmunization and recent advances in the understanding thereof.
Learning Objectives
To understand how the different cellular players of the humoral immune system work together and interact to regulate immune responses to antigens on transfused RBCs
To understand how transfused RBCs differ as an antigen from other immune stimuli
To review recent advances in understanding of cellular mechanisms of RBC alloimmunization
Introduction
William of Ockham was a medieval scholar who is attributed with the notion that the simplest explanation is the most likely, or at the very least, all other things being equal, the simplest hypothesis is more likely to be correct. However, it seems very obvious that William never encountered the cellular basis for how the immune system responds to antigens in general or to alloantigens on red blood cells (RBCs) in particular. It has been known for over a century that when a person is exposed to a foreign antigen (a body), the immune system can generate a specific response that attacks the body (eg, “antibody”). Antibodies, produced by B-cell lymphocytes, were proposed to fit like a lock and key, with a specific antibody recognizing a unique protein. Given that the number of antigens the immune system is likely to encounter is potentially limitless, B-cell diversity (thereby leading to antibody diversity) is achieved through genetic recombination of their immunoglobulin genes. In this context, the simplest explanation is that when a foreign body is encountered by the immune system, B cells whose immunoglobulins recognize the foreign body become activated and secrete relevant antibodies. Simple, right? In actuality, the cellular processes that regulate humoral alloimmunization are quite complicated, involving multiple cell types (and subtypes) in different states of maturation and/or activation, the function of which change as they traffic through different anatomical locations and microenvironments. The response is regulated by genetics, the environment, and the stochastic nature of immune biology.
General cellular paradigm of humoral alloimmunization
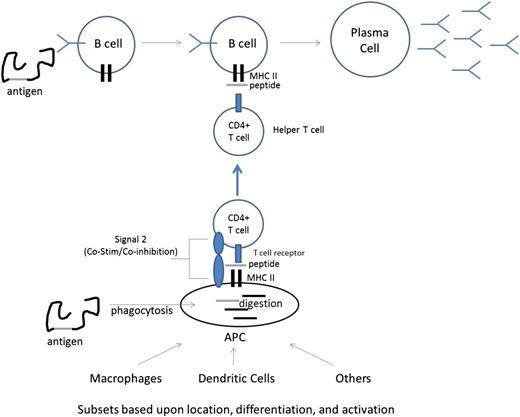
In actuality, the process that leads to a particular B cell being activated and secreting the antibody it encodes (eg, becoming a plasma cell) is complex, requiring multiple cellular and molecular players (see Figure 1 for an overview). When a foreign antigen is encountered by the immune system, at least 2 events simultaneously occur. First, antigen-presenting cells (APCs) are constantly sampling their environment through ongoing phagocytosis and can influence both the specificity and the magnitude of an immune response. Once an antigen/protein/foreign body is phagocytosed by an APC, proteins are broken down into peptides, which are then loaded into the cleft of major histocompatibility complex II (MHCII) molecules and are presented on the surface of the APC. Second, B cells consume antigen as a function of their B-cell receptor (BCR), which is a membrane-bound immunoglobulin, and can be later converted into a secreted form if the B cell becomes a plasma cell. Because B cells consume antigen via receptor-mediated endocytosis (using their BCR), the B cells selectively consume antigen recognized by their particular rearranged immunoglobulin. Similar to APCs, B cells process the antigen and present it as peptides loaded into MHCII (in some cases, B cells are themselves classified as APCs for this reason). It is worth noting that BCR-mediated endocytosis is not entirely exclusive, as it will also consume other entities that are physically associated with the target recognized by the BCR (important for linked recognition, see below); nevertheless, it is far more specific than the general consumption of antigen by other APC subsets. Thus, during RBC alloimmunization, and in the case of transfused RBCs, APCs will consume RBCs at random and present peptides on MHCII, while B cells that have a BCR specific for alloantigen on the RBCs will consume the RBC alloantigen (and possible linked material) and likewise present the same peptides on MHCII.
CD4+ T cells are similar to B cells in that they have a recombinant receptor (T-cell receptor [TCR]) that is rearranged at the genetic level, resulting in different TCRs with a distinct specificity for every naive T cell. The TCR on CD4+ T cells recognizes complexes of peptides presented in MHCII (peptide/MHCII); each TCR has specificity for a particular peptide/MHCII complex. Thus, in the case of immune responses to RBC alloantigens encountered during transfusion, recipient CD4+ T cells respond to peptides derived from donor RBC proteins, which have been processed by recipient cells and presented on MHCII.
Naive T cells that recognize an MHCII/peptide complex (through their particular TCR) on an APC can engage a TCR-specific signaling cascade (called signal 1). At this point, the T cell ceases to be “naive,” as it has encountered its “cognate antigen” in the form of the MHCII/peptide complex to which it binds. While signal 1 is necessary for T-cell differentiation into a helper T cell, signal 1 is not sufficient to fully activate a naive CD4+ T cell; the eventual fate of the T cell is affected by a series of additional ligand/receptor pairs that are expressed on the APC and T cell, respectively (collectively called signal 2). Signal 2 consists of ligand receptors pairs that can signal T cells to differentiate into effector CD4+ T cells capable of “helping” B cells (in the case of costimulatory signals) or can signal T cells to a nonhelper phenotype, such as regulatory T cells, anergic T cells, or apoptosis (in the case of co-inhibitory signals). Moreover, secreted cytokines and additional regulatory factors (from a variety of cellular origins) may form an immune milieu that can affect T-cell differentiation (signal 3). Whether a T cell encounters signal 1 in the context of stimulatory or inhibitory signal 2 and how such signal 2 influences T-cell development are functions of both genetic predisposition and environmental factors. With regard to the latter, the general paradigm is that inflammation and/or tissue damage tends to favor immune activation, whereas quiescent and healthy states tend to favor lack of immune activation. In many cases, the presence of chemical moieties associated with microbial infection or abnormal tissue damage (eg, necrotic tissue death) leads to an activating signal 2. It is in this context that significant evidence has now accumulated, in both animal models and human studies, that recipient inflammation and/or immune activation at time of transfusion regulates alloimmunization to antigens on transfused RBCs.1-6
Ability of cells of the immune system to detect RBC alloantigens
It is important to note that antibody responses to alloantigens on RBCs represent the recognition of very subtle antigenic differences. The vast majority of RBC alloantigens (other than ABO carbohydrates) consist of proteins that differ by a single amino acid polymorphism between donor and recipient (eg, K vs k). RhD is a notable exception whereby discrepancies between donors and recipients involve the whole protein (although there is extensive homology between RhD and RhCE). Moreover, RhD can exist as multiple recombined and mosaic forms so as to actually represent a family of antigens.7 This likely accounts for the observation that RhD is much more immunogenic than other RBC alloantigens; RhD-negative recipients that receive an RhD-positive RBC transfusion generate alloantibodies at a rate of 30% to 50%. In contrast, alloimmunization rates to other non–RhD-based alloantigens are only 3% to 10%, despite multiple exposures.
The MHCII gene (contained within the HLA) is highly variable from person to person. Each different MHCII has a peptide-binding pocket that can accommodate different peptides. Thus, each person has a different complement of foreign peptides that their particular MHCII can present to T cells. Given that most RBC alloantigens differ by a single amino acid, it has been hypothesized that whether or not a person becomes alloimmunized is a function of their particular HLA type. Although there are certainly data to support the notion of HLA restriction for some blood group alloantigens, such is not the case for other RBC alloantigens.8 Moreover, even for those RBC alloantigens that are HLA restricted, the correct HLA is necessary, but not sufficient; in other words, a significant percentage of transfusion recipients with the correct HLA type still do not make an alloantibody response after exposure to a particular RBC alloantigen. It is for this reason that what begets RBC alloimmunization has been attributed to other immunogenic or environmental factors, centered mostly on factors that regulate signals 2 and 3.
APC biology in RBC alloimmunization
Historically, macrophages (MØs) and dendritic cells (DCs) have been considered the predominant professional APCs, meaning they can drive activation of naive T cells at the beginning of an immune response. However, in recent decades, DCs have been considered to be the primary professional APC, with less emphasis on MØs. Nevertheless, MØs appear to have potent APC function in some settings. In some cases, B cells and other cell types stimulated to express MHCII by inflammation can also function as professional APCs. Neither MØs nor DCs are a single entity; rather, there are very important subsets of both types. MØs can exist in several different phenotypes, including M1 and M2 states, which lead to different effects upon CD4+ T-cell activation. Several distinct DC subtypes also exist, including myeloid DCs, lymphoid DCs, and plasmacytoid DCs. In actuality, the complexity of MØs and DCs goes much deeper than these categories, including differences based upon anatomical location, state of differentiation, and nature of activation.9 However, for the purposes of the current work, simpler categories will be used with relevant references for the reader interested in additional details.
Recent advances in animal modeling of RBC alloimmunization have allowed the initial dissection of the role that different APC subsets play in the initiation of RBC alloimmunization. Murine models of RBC alloimmunization have clearly shown that macrophages are predominantly responsible for the consumption of transfused RBCs upon first exposure and prior to the formation of any alloantibody response.10,11 However, RBC consumption by DCs is also detectable in these systems, albeit at a low level.12 Not only do macrophages consume more RBCs (on a per cell basis) than DCs, but also the overall number of macrophages far exceeds the number of DCs. However, although macrophages consume more RBCs than DCs, sorting experiments followed by culture with T cells specific for peptides from RBC alloantigens presented on MHCII demonstrate T-cell activation and proliferation in response to DCs that have consumed RBCs and not in response to macrophages. Similarly, while macrophages are responsible for the vast majority of clearance of stored RBCs, it appears to be the DCs that drive the CD4+ T-cell response.13 The dependency on DCs to initiate alloimmunity to RBCs has been further dissected in studies that demonstrate CD8-33D1+ DCs (bridging channel DCs) are required; in contrast, CD8+ DCs do not appear to play an essential role.14
CD4+ T-cell biology in RBC alloimmunization
As with DCs, CD4+ T cells have multiple different phenotypes and subsets, both through development and differentiation. Human studies have provided clear evidence of circulating CD4+ T cells, specific for peptides from RBC alloantigens presented by MHCII, in patients who have become alloimmunized by transfusion.15,16 T-cell restimulation assays have shown clear evidence of CD4+ T cells in response to peptides, at least in the RhD and Kell systems. Murine models have allowed a more detailed mechanistic analysis of CD4+ T cells activated in response to RBC alloimmunization, and in particular, they provide insight into functional roles and requirements. Follicular helper T cells (a particular subset of CD4+ T cells) are required for humoral alloimmunization to occur in mice. More specifically, deletion of the interleukin-6 (IL-6) receptor on CD4+ T cells prevents RBC alloimmunization, whereas IL-6 receptor deletion on B cells does not prevent antibody response.17 While much remains to be studied regarding the specifics of the role that CD4+ T cells play in RBC alloimmunization, they appear to be required (at least for some antigens) as follicular helper cells, for which IL-6 signaling is an essential pathway.
Functional effects of the different nature of B- and T-cell epitopes
There are functional implications that arise from the understanding that epitopes recognized by antibodies and epitopes recognized by CD4+ T cells are fundamentally different. Antibodies recognize native 3-dimensional structures, whereas T cells recognize linear peptides presented by an MHCII molecule. Thus, even in the case that a single amino acid polymorphism is responsible for an RBC alloantigen, the nature of the antigen seen by a T cell is different than that seen be a B cell. If the immune system encounters a linear T-cell epitope in the absence of the native antibody target, then one can be immunized in the CD4+ T-cell compartment without making a detectable antibody response. The event would not be picked up by any of the existing clinical assays in blood banking, which are all antibody based. However, should such a CD4+ T-cell–based immunization occur, it can prime a recipient for a robust alloantibody response upon subsequent exposure to a RBC expressing the alloantigen in question. This may occur if a person encounters a microbial antigen that expresses a peptide that shares the amino acid sequence of a T-cell epitope from a blood group antigen (just 12-15 amino acids), even if the rest of the molecule that carries the blood group antigen is absent. It has been demonstrated in animal models that exposure to a microbe mimicking the peptide that constitutes an RBC alloantigen does not in and of itself result in a detectable antibody response to the authentic RBC alloantigen, but it does result in a rapid and robust humoral response if RBCs expressing that alloantigen are ever transfused.18 In contrast, at least in theory, exposure to such peptides by pathways that typically beget immune tolerance (eg, orthology in other self-genes or exposure to food) may decrease rates and magnitude of alloimmunization. There is direct evidence of such pathways in humanized murine models, as demonstrated by induction of immunological tolerance through exposure to peptides, in the absence of the authentic native antigen.19 Moreover, it has been noted that a number of known microbes have amino acid sequences highly similar to peptides that contain the variant amino acid known to define serological blood group antigens.18 Thus, this remains an area of theoretical and practical importance in understanding human RBC alloimmunization.
Linked recognition of CD4+ T-cell and B-cell responses
One of the nuanced effects of the linkage of CD4+ helper T cells (that recognize peptide/MHCII complexes) and B cells that recognize native antigens is that the peptide epitope that stimulates the T cell need only be physically linked to the B-cell epitope that is recognized by the BCR on a B cell. This system “often called linked recognition” was first identified in murine models and is also observed in human immune responses. The implications of this are significant. First, it expands potential alloimmune barriers in RBC alloimmunization to polymorphisms other than those that define the blood group antigens per se. Blood group antigens are identified by those epitopes against which humans have become alloimmunized. Polymorphisms or posttranslational modifications change a 3-dimensional structure, resulting in an “epitope” that an antibody can recognize. In the case of many blood group antigens, this has been defined as a single amino acid polymorphism. It is important to note there is an observation bias in that any polymorphism that does not result in 3-dimensional change that is recognized by an immunoglobulin will never be detected by immunohematology labs, as no antisera will ever be detected in RBC crossmatch and/or screening assays. However, this does not mean that such polymorphisms do not exist; indeed, it has been reported that numerous “nonexofacial polymorphisms” (NEPs) exist in the molecules that carry the serologically detected blood group antigens.20 It has been experimentally demonstrated in mice that such NEPs can serve as CD4+ T-cell antigens that provide help to B cells recognizing a serologically defined blood group antigen, even if the NEP is distant (in the molecule) from the change that the antibody is recognizing.18 More importantly, it has been directly demonstrated in human alloimmunization to RhD that many of the dominant epitopes recognized by CD4+ T cells are in the transmembrane or cytosolic domains of RhD, meeting the definition of NEP (although allosteric conformational changes cannot be ruled out).16
The practical ramifications of NEPs and linked recognition are twofold. First, while existing blood group antigen genomics platforms (using defined single-nucleotide polymorphisms) will allow prediction of RBC phenotype, they will not pick up NEPs. NEPs could be built into the single-nucleotide polymorphism array as they are uncovered, but full sequencing of the molecules carrying the blood group antigens will be required in order to test for the presence of NEPs, as they can exist anywhere in the molecule. In the event that “NEP mismatch” affects the immunogenicity of a blood group antigen in a given recipient, and in the cases of multiply transfused patients for whom alloantibodies are a problem, then matching of NEPs may allow a decrease in RBC alloimmunization.20 Moreover, transfusion induced autoantibodies to RBC antigens has been described, and NEPs represent a mechanistic explanation for such autoantibodies.20 Matching of NEPs may also help avoid autoimmunization. Finally, as is explained in the section in linked recognition above, to the extent that microbes express peptides similar to NEPs, exposure to them they may affect RBC alloimmunization upon subsequent transfusion with RBCs expressing the entire RBC alloantigen. The extent to which NEPs play such a role is unclear, and even if they did play a role, it is unknown if the added benefit of matching NEPs would justify the resources to do so; however, it is a biological implication of linked recognition in the immune response and should be kept within our thinking.
B-cell biology in RBC alloimmunization
B cells are the ultimate source of antibodies, which are made when B cells differentiate into plasma cells that secrete immunoglobulins. As indicated above, B cells typically require CD4+ T-cell help; nevertheless, B cells are the synthetic source of secreted immunoglobulins. It is worth noting that many subsets of B cells are now recognized, based upon both phenotype and anatomical location. Some B cells are appreciated to have immunoregulatory roles, irrespective of the immunoglobulin they encode.21 Moreover, in some cases, antibody responses are CD4+ T-cell independent. Although the biochemical nature of human antibodies to RBC antigens has been carried out and a number of monoclonal human anti-RBC antibodies have been isolated, there has been no detailed analysis of where and how B cells become activated and differentiate to RBC antigens. Some animal studies have been carried out, which indicate that marginal zone B cells are essential for RBC alloimmunization.22 Marginal zone B cells are a special subset of B cells both by phenotype, and also with regards to their anatomical location, in the specialized marginal zone of the splenic architecture. As marginal zone B cells play an important role with respect to circulating particulate antigen, which RBCs can be viewed as, their importance in RBC alloimmunization would be consistent with known biology. Both mice and humans have marginal zone B cells, although they certainly differ in several regards. Moreover, human and murine splenic architecture differs in several important regards, despite also having much similarity. Nevertheless, the murine data implicate marginal zone B cells as a potential central player in RBC alloimmunization.
In addition to the requirement for marginal zone B cells in murine studies of RBC alloimmunization, the spleen itself plays an important role in RBC alloimmunization in mice, as splenectomy decreases (if not abrogates) RBC alloimmunization.3 Whether this translates to humans is a controversial issue. Many have pointed to the high rates of alloimmunization in patients with sickle cell disease (SCD) as evidence that a spleen is not required for RBC alloimmunization in humans, as many SCD patients autoinfarct their spleens and are seen as functionally asplenic. However, despite autosplenectomy of SCD patients, their lymphatic white matter persists, and spleens essentially grow back in response to therapeutic intervention (eg, hydroxyurea). Moreover, the murine data report that a spleen is required for initial alloimmunization and do not address responses that occur in a splenectomized individual who was previously exposed to RBC alloantigens when a spleen was present. Thus, the extent to which the spleen is the actual site of RBC alloimmunization or is required for such immunization in humans remains unclear, as it has not yet been thoroughly investigated.
Conclusion
Over the last century, careful and applied serology has identified in excess of 340 different RBC alloantigens. Studies of human transfusion recipients have elucidated the nature of these RBC alloantigens, their immunogenicity, and the clinical significance of alloantibodies against them. Meanwhile, basic and translational immunology has defined and elucidated the fundamental workings of humoral immune responses. Although not yet completely understood, the basic cellular and molecular paradigms of humoral immune response predict much of observable human and animal immunology. Alloimmunization to transfused RBCs has characteristics that are quite distinct from better-studied immune responses (eg, vaccines and microbial infections). Transfused RBCs are a sterile source of antigen, and while they may not be without “danger signals,” such would not be of microbial origin. They represent a very large dose of antigen that is given intravenously and persists in the circulation for long periods. Except in the case of injury, RBCs are confined to the circulation and do not typically enter the lymphatics, thus being excluded from a large part of the immune system. Finally, the antigenic differences between donor and recipient RBCs are very small compared with an entirely foreign microbe. Indeed, this constellation of characteristics results in a weakly immunogenic stimulus that fails to induce immunity in the majority of recipients and likely causes tolerance. The journey to find out how the particulars of alloimmunization to RBCs fit into the cellular and molecular mechanisms of the humoral immune system has just begun. Recent advances are detailed above, and ongoing studies have the potential to continue to elucidate our understanding of cellular immunology of RBC alloimmunization. Given the particulars of RBC alloimmunization, it remains as likely that the study of transfusion will uncover as much novel understanding of immunology in general as the application of immunology will elucidate particulars of RBC alloimmunization.
Correspondence
James C. Zimring, BloodworksNW Research Institute, 1551 Eastlake Ave E, Suite 100, Seattle, WA 98102; e-mail: jzimring@bloodworksnw.org.
References
Competing Interests
Conflict-of-interest disclosure: J.C.Z. is on the board of directors and an advisory committee for Rubius Therapeutics and has received research funding from Immucor Inc. K.E.H. declares no competing financial interests.
Author notes
Off-label drug use: None disclosed.