Abstract
Acute myeloid leukemia (AML) is a disease with marked heterogeneity in both response to therapy and survival. Cytogenetics, age, and performance status have long determined prognosis and therapy. The advent of molecular diagnostics has heralded an explosion in new prognostic factors, including gene mutations in KIT, FLT3 (Fms-like tyrosine kinase 3), NPM1 (nucleophosmin 1), and CEBPA (CCAAT enhancer-binding protein-α). Microarray technology can now identify unique gene expression signatures associated with prognosis. Similarly microRNA expression, single nucleotide polymorphism arrays, and DNA methylation signatures have recently described important new prognostic subgroups of AML, and are contributing to our understanding of AML disease biology. Combined with proteomic profiling, these technologies have helped identify new targets and signaling pathways, and may soon help to identify individual patients likely to benefit from specific therapies, including allogeneic hematopoietic cell transplantation. In summary, new clinical and molecular prognostic markers have begun to significantly improve our understanding of AML biology. We are now close to a time when we will be able to use these prognostic factors and technologies to identify new targets for therapy and to determine who may benefit from that therapy, and ultimately change how we treat individual patients with AML.
It has long been appreciated that acute myeloid leukemia (AML) is a clinically heterogeneous disease, with marked differences in survival following intensive chemotherapy based on age,1 blast-cell morphology, and cytogenetic abnormalities.2,3 Although therapeutic advances have lagged over the past two decades, there has recently been an explosion in new prognostic factors in AML, which is driving our understanding of disease biology and also the development of new therapeutic targets. This is particularly the case in patients with normal cytogenetics, who comprise the largest subgroup of AML patients (approximately 45%),2,4 where many new important prognostic factors have been identified, including gene mutations in FLT3 (Fms-like tyrosine kinase 3; generally unfavorable), NPM1 (nucleophosmin 1), and CEBPA (CCAAT enhancer-binding protein-α; generally favorable). Clinical and genetic prognostic markers are now central in the evaluation of AML patients and are critical in guiding rational management in the clinic, including selection for targeted clinical trials (eg, FLT3 tyrosine kinase inhibitors for those with FLT3 mutation) or consolidation of remission with allogeneic transplantation. Indeed, AML is increasingly subclassified as unique disease entities in the 2008 revision of the World Health Organization classification of myeloid neoplasms and acute leukemia3 based on specific recurring genetic abnormalities that predict prognosis and response to therapy. With the exception of some of these specific disease entities, there has been a move beyond individual gene mutations toward integrating clinical and genetic prognostic data more broadly in determining prognosis and therapy. This review will address new clinical, genetic, and phenotypic prognostic markers, with an emphasis on the evaluation of patients with cytogenetically normal (CN) AML.
It is important to emphasize that prognostic factors are highly dependent on the available therapies. In many cases (particularly for younger AML patients), the rational application of prognostic markers may allow a decision in postremission therapy between high-dose cytarabine versus allogeneic hematopoietic cell transplantation (HCT). The adverse prognostic impact of some genetic features—such as poor-risk cytogenetics, high EVI1 expression, or an internal tandem duplication (ITD) mutation in FLT3 (FLT3-ITD) among others—may be abrogated at least in part by allogeneic HCT.5–7 Intriguingly, some prognostic markers are also potential targets of therapy, such as tyrosine kinase inhibitors targeting mutant FLT38 or the use of small molecules or monoclonal antibodies targeting the chemokine receptor CXCR4, the overexpression of which is associated with inferior prognosis. Efforts to target P-glycoprotein (P-gp; the MDR1 gene product, long associated with worse prognosis through chemoresistance) have unfortunately failed to show improvements in survival. However P-gp remains a relevant prognostic marker for future trials of chemotherapeutic agents that are not substrates for P-gp–mediated drug efflux.
A recent meta-analysis of allogeneic HCT in first complete remission (CR), including over 6000 patients in 24 prospective trials, demonstrated a significantly lower hazard ratio (HR) of death for both intermediate-risk (HR 0.83; 95% CI, 0.74–0.93) and high-risk (HR 0.73; 95% CI, 0.59–0.90) cytogenetic subgroups, compared with nonallogeneic HCT strategies.5 In a similar study, comparing patients in CR with versus without a donor, it has been estimated that the beneficial effect of allogeneic HCT becomes apparent as soon as the risk of relapse exceeds approximately 35%, irrespective of the specific underlying cytogenetic abnormality that causes the higher relapse rate.9 Furthermore, retrospective studies suggest that allogeneic HCT may negate the prognostic impact of FLT3-ITD and unmutated NPM1.6 Although this makes allogeneic HCT an attractive alternative in this setting, prospective studies are necessary to confirm its potential benefit with newer molecular prognostic factors.
Refinements in Cytogenetics and Prognosis in AML
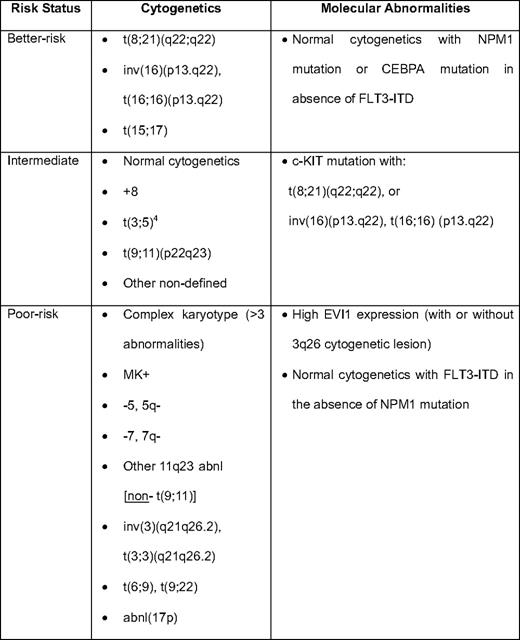
Cytogenetics remain the most important disease-related prognostic factor in AML, and are well-characterized as “favorable,” intermediate risk, and adverse (Table 1).2,10 This has been reviewed extensively in Hematology 2009.11 Those with “intermediate” prognosis cytogenetics represent the largest and most heterogeneous group of patients, and, as outlined below, there have been great efforts to identify those in this group who are at higher risk and who may be candidates for allogeneic HCT. The availability of large national databases with long-term follow-up has allowed some refinements to the cytogenetic classification, including the identification of rare abnormalities (< 1%) with prognostic impact (eg, isolated t(3;5) associated with NPM1-MLF1 fusion, which occurs at younger age and has an intermediate prognosis).4 A recent meta-analysis from the German AML Intergroup study has further strengthened the observation of significantly better prognosis among patients with isolated t(9;11) relative to other translocations involving 11q23 (particularly t(6;11), which is associated with a very poor survival and does not appear to benefit from allogeneic HCT12 ).
An important recent observation from the Hemato-Oncologie voor Volwassenen Nederland (HOVON) group is the existence within the adverse cytogenetic risk AML of the so-called “monosomal karyotype” (MK+), characterized by the presence of an autosomal monosomy in conjunction with at least one other autosomal monosomy or structural abnormality.13 Patients with MK+ have a dismal prognosis, with ≤ 5% long-term survival, and the prognostic impact of MK+ has been confirmed in secondary AML14 and by other groups.4
Another significantly adverse genetic subgroup is high expression of EVI1, most commonly in conjunction with lesions on chromosome 3q26, which—although relatively uncommon (≤ 10% of new patients)—is associated with an aggressive form of AML with very poor prognosis.7 Because of the possible benefit from allogeneic transplantation in the setting of high EVI1 expression (albeit based on a relatively small number of reported patients), it has been suggested that pretreatment EVI1 screening should be included in risk stratification.7
Gene Mutations in AML
Prognostically important gene mutations have been identified in all subtypes of AML and contribute to disease progression. The most prognostically important and prevalent mutations in different cytogenetic subgroups are reviewed below.
KIT Mutations in Core-Binding Factor Leukemia
Although patients with AML and inv(16)(p13.q22) and t(8;21)(q22;q22)—so-called “core-binding factor” (CBF) leukemia—in general have a more favorable prognosis, there remains a significant failure rate, and the long-term disease-free survival is only approximately 60%.2,4 Retrospective cooperative group studies have demonstrated activating KIT mutations in approximately 30% to 40% of patients with inv(16) associated with significantly higher incidence of relapse and significantly lower survival.15 In those with t(8;21), the incidence of KIT mutation (and, in particular, D816 in the KIT tyrosine kinase domain) appears to be more variable (overall approximately 20%–30%), and there was a similar adverse effect on relapse and survival,16 although in a separate CALGB (Cancer and Leukemia Group B) study, there was not an independent effect of KIT mutation on survival.15 KIT mutation testing, therefore, appears to be prognostically important for survival in CBF AML and has recently been incorporated into the National Comprehensive Cancer Network guidelines.17 Whether patients with KIT mutations preferentially benefit from allogeneic HCT is not known.10
Gene Mutations in CN-AML
CN-AML is the largest cytogenetic subgroup of the disease and is marked by heterogeneous survival. Gene mutations studies can help identify genetic lesions associated with treatment failure, and a number of occult gene mutations have been described that have significant prognostic impact and allows discrimination of patients who may be candidates for alternative therapies, including allogeneic HCT.
FLT3 Mutations
FLT3 is an important class III receptor tyrosine kinase normally expressed in early bone marrow progenitors, where it plays an important role in hematopoiesis. Mutations in FLT3 are among the most prevalent genetics lesions in AML, most commonly an ITD in the FLT3 juxtamembrane domain that occurs in approximately 25% of adult AML patients.18 Patients with a FLT3-ITD often have a higher white blood cell count and typically present with CN-AML, where almost one-third of patients harbor an occult FLT3 mutation.6 Early retrospective studies demonstrated that the FLT3-ITD mutation is consistently associated with significantly worse survival in “Younger” adults (age < 60 years) (reviewed in Gilliland and Griffin18 ). The incidence of FLT3 mutations increases with age, but the FLT3-ITD appears to have less prognostic impact among “older adults” (age > 60 years),19 possibly because other adverse prognostic factors are more prevalent.
Mutations in the FLT3 tyrosine kinase domain (TKD) occur in approximately 5% to 7% of adult AML patients, also most commonly in CN-AML, so that, overall, one-third of AML patients will have an occult FLT3 mutation (either ITD or TKD). Unlike FLT3 ITD mutations, the presence of a TKD mutation does not in general appear to carry an independent adverse prognosis,20 although some evidence suggests that those with a higher FLT3-TKD mutant level may indeed have an inferior disease-free survival.
Patients with a FLT3-ITD do not appear to benefit from anthracycline dose intensification,21 but may benefit from allogeneic HCT.6 Tyrosine kinase inhibitors targeting FLT3-ITD have shown clinical activity in phase I studies8 (albeit with very few complete remissions) and a large CALGB-led international multigroup phase III trial of the FLT3 inhibitor midostaurin in combination with standard daunorubicin and cytarabine is actively accruing in patients with FLT3 mutation (CALGB 10603).
Mutations in NPM1
The discovery of mutations in the NPM1 gene in AML was based on the fortuitous observation of aberrant cytoplasmic (rather than nuclear) localization of NPM identified by immunohistochemistry.22 NPM is nucleocytoplasmic shuttling protein mainly localized in the nucleolus that has multiple functions: (a) it interacts with p53 in controlling cell proliferation and apoptosis, (b) is involved in the maintenance of genomic stability controlling DNA repair and centrosome duplication during mitosis, and (c) plays a key role in ribosome biogenesis. The NPM1 gene belongs to a new category that functions both as an oncogene and tumor-suppressor gene, depending on gene dosage, expression levels, interacting partners, and compartmentalization.
NPM1 mutations are now recognized as the most common genetic lesion in AML, present in 50% to 60% of patients with CN-AML, typically with M4 blast cell morphology, a female preponderance, usually lacking in expression of CD34, and with a distinct gene expression profile.22 The gene and microRNA expression profiles suggest that NPM1 mutations define a biologically homogeneous entity in CN-AML, and AML with mutated NPM1 represents a unique genetic lesion23 that is now recognized as a provisional entity in the 2008 revision of the World Health Organization classification of myeloid neoplasms and acute leukemia.3
NPM1 mutations are prognostically favorable in the absence of FLT3-ITD.6 NPM1 mutations also have favorable prognostic impact in older patients with CN-AML, especially those age ≥70 years.
CEBPA Mutations
CEBPA encodes for CCAAT/enhancer binding protein-α, a granulocytic differentiation factor important in the regulation of myeloid progenitors. Mutations in CEBPA occur in approximately 5% to 10% of de novo AML and is most common in CN-AML.6 A germline CEBPA mutation has also been reported in familial AML.24 In the absence of a FLT3-ITD, a CEBPA mutation confers significantly better prognosis in patients with CN-AML, with approximately 60% long-term survival, particularly if there is a “double” (biallelic) mutation that occurs in one-half to one-third of those harboring a CEBPA mutation.25 In one study, the presence of a CEBPA mutation was independently associated with better survival in CN-AML, even in the presence of other high-risk molecular features, such as FLT3-ITD.26
RAS Mutations in AML
Mutations in NRAS and KRAS occur in approximately 10% and 5% of AML patients, respectively, and may indeed be more common than previously appreciated.27 RAS mutations only rarely occur in conjunction with FLT3 mutation and do not appear to have a significant impact on AML survival. A recent study suggests that patients with CN-AML and RAS mutations may benefit from high-dose cytarabine consolidation.28
Other Gene Mutations With Potential Prognostic Importance
Mutations in the isocitrate dehydrogenase 1 gene occur in approximately 10% to 15% of CN-AML and may have a significant adverse impact on survival, especially among those with other higher risk features (unmutated NPM/FLT-ITD).29 Isocitrate dehydrogenase 2 mutations have also recently been described in CN-AML and contribute to a lower CR rate.30 TET2 mutations occur in up to 25% of patients with secondary AML, although the impact on survival is not known.31 Partial tandem duplication in the MLL gene (MLL-PTD) has been reported in approximately 5% of AML patients, particularly those with therapy-related secondary AML. It is, in general, associated with a poor prognosis, although a more recent study from CALGB suggested that better long-term outcomes may be achieved with intensive consolidation, including autologous stem cell transplantation.32 High expression of Id1 (“inhibitors of differentiation”), which occurs more frequently with FLT3-ITD, is independently associated with significantly lower CR rates and overall survival in CN-AML.33 Not surprisingly, mutations in TP53 have been reported in AML with complex karyotype and are associated with very poor prognosis.19,34
Integrating Gene Mutations Into Clinical Management
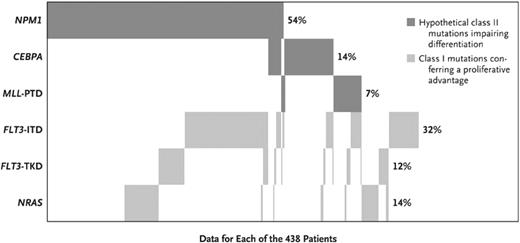
Although many occult gene mutations have been reported in CN-AML, the most common are NPM1 mutations and FLT3-ITD. However, there is significant overlap between these two mutations. The impact on clinical outcome in the setting of a normal karyotype was recently demonstrated in a large retrospective analysis (n = 872) of CN-AML patients younger than age 60 treated on prospective trials of the German-Austrian AML Study Group that included allogeneic HCT.6 Patient samples were tested for mutational status of FLT3-ITD (31% positive), FLT3-TKD (11% positive), NPM1 (53% positive), CEBPA (13% positive), MLL-PTD (7% positive), and NRAS (13% positive). Importantly, approximately one-third of patients harboring an NPM1 mutation also had an FLT3-ITD (Figure 1). The risk of relapse or death was significantly lower in patients with an NPM1 mutation without FLT3-ITD (HR 0.44; 95% CI, 0.32–0.61) and those with CEBPA mutation (HR 0.48; 95% CI, 0.30–0.75). Patients not in these two more favorable categories (including those with an FLT3-ITD) had a significantly worse outcome. The adverse impact of FLT3-ITD appeared to be negated by allogeneic HCT in this study, although patients were not selected for transplantation on that basis, thus raising the potential concern of patient selection. Nevertheless, FLT-ITD patients, or those without the more favorable CEBPA mutation or NPM1 mutation without FLT3-ITD, represent a high-risk group of patients with CN-AML.
Frequencies and distribution of the NPM1, CEBPA, MLL, FLT3, and NRAS mutations, according to mutation class. Frequencies are given for the 438 patients in whom data on all genes were available. PTD indicates partial tandem duplication. Reprinted with permission from the New England Journal of Medicine.6
Frequencies and distribution of the NPM1, CEBPA, MLL, FLT3, and NRAS mutations, according to mutation class. Frequencies are given for the 438 patients in whom data on all genes were available. PTD indicates partial tandem duplication. Reprinted with permission from the New England Journal of Medicine.6
The incorporation of gene mutation studies for these genetic abnormalities—and in particular CEBPA, FLT3-ITD, and NPM1 mutations in CN-AML (and KIT mutations in CBF leukemia)—allows the distinction of prognostically unique subgroups (Figure 2; Tables 1 and 2), and is now an available and appropriate (albeit still optional) test in the initial workup of a patient with AML.10,17
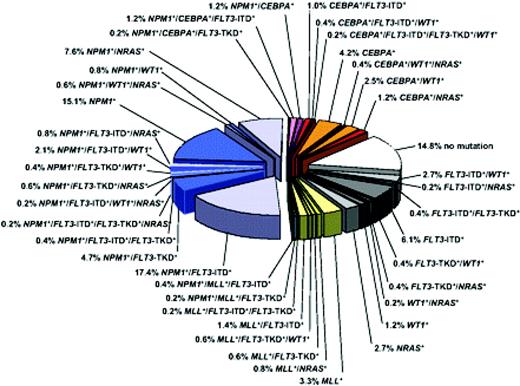
Molecular heterogeneity of CN-AML based on mutations in the NPM1, CEBPA, MLL, FLT3 (ITD and TKD mutations at codons D835 and I836), NRAS, and WT1 genes. Blue colors denote NPM1-mutated subsets, orange/red colors denote CEBPA-mutated subsets, and yellow/green colors denote MLL-mutated subsets. Gray colors depict subsets without hypothetical class II mutations, and the white sector shows the subset without any mutation in the previously described genes. Data are derived from mutational analysis of 485 younger adult patients with CN-AML from the German AML Study Group. Reprinted with permission from Blood.10
Molecular heterogeneity of CN-AML based on mutations in the NPM1, CEBPA, MLL, FLT3 (ITD and TKD mutations at codons D835 and I836), NRAS, and WT1 genes. Blue colors denote NPM1-mutated subsets, orange/red colors denote CEBPA-mutated subsets, and yellow/green colors denote MLL-mutated subsets. Gray colors depict subsets without hypothetical class II mutations, and the white sector shows the subset without any mutation in the previously described genes. Data are derived from mutational analysis of 485 younger adult patients with CN-AML from the German AML Study Group. Reprinted with permission from Blood.10
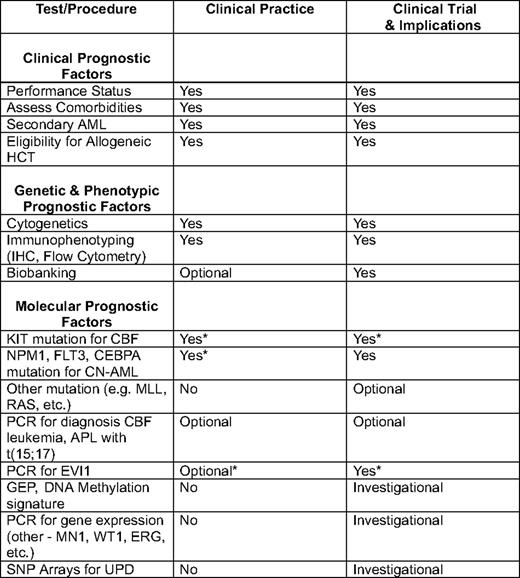
IHC indicates immunohistochemistry; PCR, polymerase chain reaction; APL, acute promyelocytic leukemia; UPD, uniparental disomy.
*Not recommended in all guidelines.10
Altered Gene Expression in AML
In addition to the presence of occult mutations, markedly increased or decreased expression of specific genes (typically those involved in hematopoiesis, myeloid differentiation, or immune response) has been significantly associated with response to therapy and with survival.
BAALC Expression
High expression levels of the brain and acute leukemia, cytoplasmic (BAALC) gene, has been associated with significantly lower CR rates and inferior survival, although BAALC expression also frequently associates with other adverse molecular prognostic features, including FLT3-ITD, lack of NPM1 expression, and high ERG (ETS-related gene) expression. Patients with high BAALC expression undergoing consolidation with allogeneic HCT were observed to have a lower cumulative incidence of relapse, suggesting a potential benefit in this adverse prognostic group.35
MN1 Expression
The meningioma 1 (MN1) gene encodes a protein that participates in a gene transcription regulator complex involving retinoid receptors. High MN1 expression has been identified as an adverse prognostic factor in CN-AML commonly associated with other adverse cytogenetic and molecular prognostic features.36
WT1 Expression
Mutations in the Wilm's tumor 1 (WT1) gene have been reported in approximately 10% of cases of CN-AML and tend to occur in younger patients with a higher white blood cell or blast count. The impact of WT1 mutations on survival remains controversial. WT1 mutation has been independently associated with significantly inferior response to therapy and significantly lower overall survival, even in the setting of FLT3-ITD or wild-type (nonmutant) NPM1 in some studies,37 whereas in other recent studies of CN-AML patients, there was no impact on survival.38 Interestingly, the presence of a known germline WT1 single nucleotide polymorphism (SNP) found in 25% of CN-AML patients in one of the latter studies was associated with significantly better survival.38 Failure to reduce WT1 transcripts below the threshold limits defined in normal controls by the end of consolidation also predicted increased relapse risk (P = .004), suggesting that WT1 may be used to monitor minimal residual disease after therapy.39
ERG Expression
ERG effects downstream signaling pathways involved in cell proliferation, differentiation, and apoptosis. High-level expression of ERG is associated with increase cumulative incidence of relapse and significantly worse survival in CN-AML, and may at least partially account for early relapses in the otherwise more favorable NPM1 mutated/FLT3 wild-type subgroup.40
Gene Expression Profiling
Although changes in the expression of individual genes are of interest and potentially prognostically important, there has been an effort to integrate gene expression globally in AML prognosis. Distinct gene expression signatures have now been identified using gene expression profiling (GEP) and have allowed identification of prognostically important subgroups of AML, in particular in CN-AML.41 Importantly, GEP studies in older AML patients have demonstrated unique signatures that, in addition to a prognostic impact, also associate with genes involved in proliferation and apoptosis, and with dysregulation of oncogenic signaling pathways, altered tumor environment, and signatures of chemotherapy sensitivity. Because many of the genetic alterations are potential targets for specific therapeutic intervention, the use of GEP (in conjunction with analysis of specific mutations, for example, FLT3) is likely to have a major impact on future clinical management, including determining sensitivity to specific therapies (eg, anthracyclines42 ), particularly in CN-AML.
The clinical utility of GEP has recently been tested in an international effort of 11 laboratories on three continents and including samples from over 3000 patients to validate whole genome expression profiles. Preliminary reports suggest that the technology is robust for the diagnosis of hematologic malignancies with high accuracy.43 The extent to which this will replace current diagnostic and prognostic markers remains to be determined, particularly in view of the rapid expansion in other profiling technologies (eg, DNA methylation arrays) and the development of accessible and cheaper whole genome mutation analyses.
MicroRNA Signatures
MicroRNAs are noncoding RNAs of 19 to 25 nucleotides in length that regulate gene expression. They perform critical functions in biological processes, including differentiation, stress response, apoptosis, and proliferation. They have been shown to play a role in malignant transformation in other malignancies, and recent studies in AML have shown that specific patterns of microRNA expression are closely associated with cytogenetics and the presence of an FLT3-ITD. In CN-AML with high-risk features (lack of NPM1 mutation or the presence of FLT3-ITD), a microRNA expression signature comprised of 12 probes was identified that was inversely associated with event-free survival.44 Provocatively, there was also an inverse correlation with the expression of gene-encoding proteins involved in innate immunity. These results indicate a relationship between microRNA expression and gene expression in CN-AML, and suggest a plausible mechanistic pathway for an aggressive AML phenotype.
SNP Arrays in AML
The use of an SNP microarray for whole genome scanning allows detection of occult cytogenetic lesions in CN-AML. In addition to detecting unbalanced cytogenetic lesions, studies with SNP arrays in CN-AML demonstrate that approximately one-third of patients have a uniparental disomy (ie, the copy-neutral loss of heterozygosity undetectable by standard cytogenetics).45 Uniparental disomy appears to be a previously unappreciated, but common genetic lesion in AML,46 and in the former study was associated with significantly worse survival in CN-AML.
Proteomic Profiling
The expression and activation (phosphorylation) of proteins involved in apoptosis, signal transduction pathways, and cell cycle regulation are of great interest in hematologic malignancy, and functional proteomic profiling has recently been studied in AML. Using reverse-phase protein arrays to distinguish a total of 51 protein and phosphoprotein epitopes, seven separate proteomic signature groups were generated that are able to distinguish between different FAB morphologic subtypes of AML and can discriminate signatures within CN-AML with FLT3-ITD with a significantly different survival.47 In addition to prognosis, this exciting technology will likely be of most value in ascertaining the activation status of critical signaling pathways in AML and may in the future help rationally select targeted therapies in individual patients.
DNA Methylation Arrays
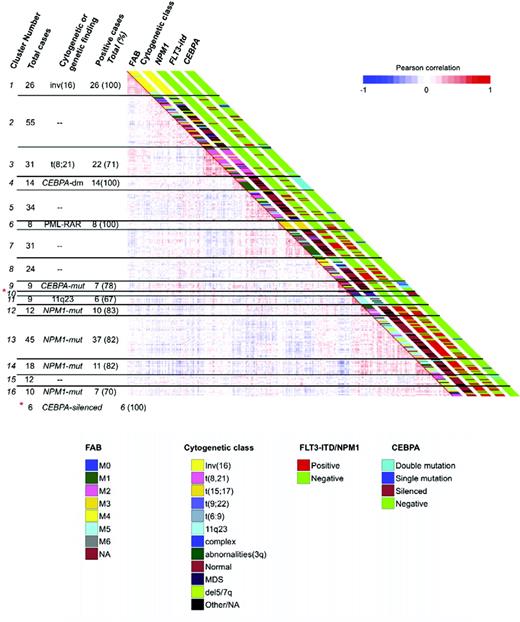
DNA cytosine methylation is a key mechanism of epigenetic regulation of gene expression, and disruption of normal DNA methylation is a hallmark of cancer. Aberrant methylation of specific genes has been described in leukemia. The fascinating development of genome-wide DNA methylation microarrays has allowed the description of novel AML subgroups independently predictive of overall survival based on DNA methylation patterns when controlled for standard established prognostic variables (eg, age, cytogenetics, and FLT3 mutational status).48 Importantly, established AML entities (eg, t(8;21), t(15;17) with PML-RARA [promyelocytic leukemia-retinoic acid receptor-α], CEBPA aberrations) were confirmed to have specific methylation profiles, but other patients with similar methylation profile and indistinguishable survival yet lacking the aberration (ie, t(8;21)-negative) were also identified (Figure 3). In addition, new AML subgroups with no other shared features have also now been described, each with deregulation in different canonical signaling pathways and with significantly different survival.48 If validated, the use of DNA methylation signatures has the potential to significantly improve our prognostic capabilities, as well as our understanding of AML disease biology.
DNA methylation segregates AML patients into 16 groups. Heatmap representation of a correlation matrix in which each patient's DNA methylation profile is correlated with that of the other patients in the data set. Patients are ordered according to the unsupervised analysis (hierarchical clustering) results, so that highly correlated patients are located next to each other. Parallel bars on the right of the heatmap have been used to indicate the principal cytogenetic and molecular findings for each patient. Cluster membership and cluster feature summaries are described on the left-hand side of the heatmap. Reprinted with permission from Cancer Cell.48
DNA methylation segregates AML patients into 16 groups. Heatmap representation of a correlation matrix in which each patient's DNA methylation profile is correlated with that of the other patients in the data set. Patients are ordered according to the unsupervised analysis (hierarchical clustering) results, so that highly correlated patients are located next to each other. Parallel bars on the right of the heatmap have been used to indicate the principal cytogenetic and molecular findings for each patient. Cluster membership and cluster feature summaries are described on the left-hand side of the heatmap. Reprinted with permission from Cancer Cell.48
Other Novel Prognostic Markers in AML
CXCR4 and VLA-4 Expression by Flow Cytometry
The chemokine receptor CXCR4 is expressed and functional in AML, and high expression by flow cytometry is an independent adverse prognostic factor in retrospective studies.49 It is potentially an attractive target in AML due to the availability of small molecules and humanized monoclonal antibodies targeting CXCR4, and clinical trials with these agents alone and in combination with chemotherapy are ongoing. The expression of “very late antigen-4” (VLA-4) on myeloblasts is not prognostic on its own, but when bound to soluble VCAM-1, its ligand, it is associated with significantly longer survival.50 This observation is of significant biological interest and suggests that VLA-4 may also be a potential therapeutic target.
“New” Clinical Prognostic Factors
In addition to well-described clinical prognostic factors (eg, age and comorbidities), socioeconomic status and gender have recently been identified as important prognostic factors, with a significantly lower survival reported for men and for “blue-collar workers” (HR 1.22; 95% CI, 1.22–1.34) in a Swedish registry study.51 Race may also have an impact, because African-American males have been found to have a significantly lower CR rate and worse survival in patients treated in CALGB studies.52 These suggest the existence of disparities in AML survival.
Although seemingly intuitive, the timing and response to initial therapy also have significant prognostic importance. Treatment delays longer than 5 days have been associated with significantly lower CR rates and worse survival in younger (but not older) AML patients.53 The quality of the CR also has prognostic importance—long-term survival is significantly less likely among patients who achieve CRp (ie, CR with incomplete platelet recovery).54 Therefore, timely initiation of therapy and proper assessment of response are imperative.
Conclusions
There have been great strides in understanding disease biology in AML through the use of new technologies, and these have brought with them the identification of novel disease-related prognostic markers that impact significantly on AML classification, response to therapy, and survival. This is particularly the case in CN-AML, where provisional new disease entities and therapeutic targets have been identified based on these studies. Although certainly most important for patients who will participate in clinical trials, testing for mutations for KIT in CBF leukemia, and for FLT3, NPM1, and CEBPA in CN-AML, are now common in clinical practice and may begin to influence treatment decisions for individual patients. It is clear that we are close to an era of individualized diagnosis—and possibly therapy—for patients with AML. As therapy for AML advances, the importance of individual prognostic factors will need to be reassessed in light of new treatments.
Disclosures
Conflict-of-interest disclosures: The author has received research funding from Ambit and Genzyme, and has consulted for Antisoma.
Off-label drugs: Use of FLT3 inhibitors in clinical development.
Correspondence
James M. Foran, MD, FRCPC, Associate Professor of Medicine, Division of Hematology/Oncology, University of Alabama at Birmingham, 1802 6th Ave. South, NP2565, Birmingham, AL 35294; Phone: (205) 934-2248; Fax: (205) 934-1608; e-mail: james.foran@ccc.uab.edu