Abstract
Hodgkin lymphoma (HL) is derived from mature B cells and subdivided into classical HL and nodular lymphocyte predominant HL (NLPHL). HL is unique among human B cell lymphomas because of the rarity of the lymphoma cells, the Hodgkin and Reed-Sternberg (HRS) cells in classical HL and the lymphocyte-predominant (LP) cells in NLPHL, which usually account for 0.1% to 10% of the cells in the affected tissues. Moreover, HRS cells are unique in the extent to which they have lost their B cell–typical gene expression pattern. Deregulation of transcription factor networks plays a key role in this reprogramming process. HRS cells show strong constitutive activity of the transcription factor NF-κB. Multiple mechanisms likely contribute to this deregulated activation, including signaling through particular receptors and genetic lesions. Inactivating mutations in the TNFAIP3 tumor suppressor gene, encoding a negative regulator of NF-κB activity, were recently identified in about 40% of patients with classical HL. HRS cells are latently infected by Epstein-Barr virus in about 40% of patients, and an important role of this virus in HL pathogenesis—in particular for cases in which HRS cells had lost the capacity to express a B-cell receptor due to destructive somatic mutation—was recently substantiated.
With an incidence of about 3 new cases per 100,000 persons per year in Western countries, Hodgkin lymphoma (HL) is one of the most frequent lymphomas. About 95% of cases belong to the classical form of the disease, while the remaining 5% represent nodular lymphocyte predominant HL (NLPHL).1 Based on differences in the histological picture and the cellular composition, classical HL is further subdivided into nodular sclerosis, mixed cellularity, lymphocyte-depleted and lymphocyte-rich forms. In classical HL, the tumor cells are designated Hodgkin and Reed-Sternberg (HRS) cells, while in NLPHL, they are now called LP (lymphocyte predominant) cells (previously they were called L&H—lymphocytic and histiocytic—cells).1 The two types of HL differ in the morphology and immunophenotype of the lymphoma cells, as well as in the composition of the cellular microenvironment, ie, the histological picture. Moreover, HRS cells likely derive from germinal center (GC) B cells that had acquired unfavorable immunoglobulin V gene mutations and normally would have undergone apoptosis, whereas LP cells derive from antigen-selected GC B cells.2 Additionally, different genetic lesions appear to be involved in the pathogenesis of these lymphomas.2 The HRS and LP cells usually account for 0.1% to 10% of the cells in the tissue, which is one of the reasons that hampered and still hampers their molecular analysis. Nevertheless, in recent years considerable progress has been made in elucidating the nature of the HRS and LP cells and the pathogenesis of HL, and key findings will be discussed in the following.
Deregulated Transcription Factor Networks and the Lost B-cell Phenotype in HRS Cells
The detection of clonally rearranged immunoglobulin V (IgV) heavy and light chain genes with many somatic mutations unequivocally showed the derivation of HRS cells from mature B cells at a GC or post GC stage of differentiation, because somatic hypermutation of IgV genes specifically takes place in antigen-activated B cells proliferating in the GC microenvironment.3 While most B-cell lymphomas retain key features of their cell of origin during malignant transformation,4 the mature B-cell origin of HRS cells was not obvious because these cells show a very unusual immunophenotype with expression of genes of many different hematopoietic cell types. Most B-cell–typical genes, including the B cell receptor (BCR), CD19, CD20, Syk and A-myb, are indeed not or only very rarely expressed by HRS cells.5,6 Thus, clarifying the mechanisms for this “reprogramming” of the HRS cells and their potential role in the pathogenesis of classical HL is important. Significant progress has recently been made in this regard. Several key transcription factors that regulate the expression of many B cell–specific genes are either not expressed in HRS cells or only at strongly reduced levels. This includes Oct-2, Pu.1, Bob1 and early B-cell factor (EBF).6–9 The helix-loop-helix transcription factors E12 and E47, which are encoded by the E2A gene, are still expressed in HRS cells, but their function is impaired by the strong expression of two of its inhibitors, ID2 and ABF1, in HRS cells.9–11 Importantly, ID2 is normally expressed in NK cells and dendritic cells, supporting development of these cells and suppressing a B-cell differentiation. Curiously, HRS cells still express Pax5, the main B-cell lineage commitment factor.12 However, many Pax5 direct target genes are not expressed. Perhaps, as many genes are regulated by the concerted action of multiple transcription factors, the absence of multiple other B-cell transcription factors is the cause for the absent or low expression of Pax5 target genes.
A further key player in the reprogramming of HRS cells is most likely Notch1, which is normally expressed in T cells but also constitutively activated in HRS cells.13 This transcription factor promotes T-cell development and inhibits B-cell development of lymphoid precursors. Notch1 downregulates the expression of the key B-cell transcription factors E2A and EBF and induces the expression of ABF1.13 As Notch1 binds to Pax5 in HL cell lines,13 it may perhaps also impair Pax5 function. Notch1 appears to be activated by its ligand Jagged1, which is expressed by cells in the HL microenvironment.13 Recently, the transcription factor STAT5 was implicated in the reprogramming of HRS cells, as expression of a constitutively active form of this factor in B cells induced many phenotypic and gene expression changes that induced an HRS cell-like phenotype of the cells.14
Molecular Features of LP Cells in NLPHL
Several findings linked the origin of LP cells in NLPHL to GC B cells.3 This included the presence of clonally rearranged and somatically mutated IgV genes in these cells, with signs of ongoing somatic hypermutation in a fraction of cases. Moreover, LP cells express typical GC B-cell markers, including BCL-6 and activation-induced cytidine deaminase, and the LP cells are found in follicular structures in association with follicular dendritic cells and GC-type T-helper cells, in this regard resembling GC.3 However, a more detailed characterization of the gene expression pattern of LP cells and of pathogenetic mechanisms in these cells was still lacking. Therefore, we performed global gene expression studies of microdissected LP cells and compared these to gene expression profiles of the main normal B-cell subsets and other mature B-cell lymphomas.15 A comparison to the other normal B cells indicated that in terms of their gene expression, LP cells resemble GC B cells but have already acquired features of memory B cells, indicating that these cells may derive from late GC B cells at the transition to memory B cells.15 LP cells show a partial loss of the B cell phenotype, although this is not as pronounced as the one seen in HRS cells of classical HL.15 In spite of the resemblance of NLPHL to follicular lymphoma in terms of the histological picture and the association of the lymphoma cells with GC structures, and in spite of the immunophenotypic similarities between follicular lymphoma B cells and LP cells, the genome-wide differential gene expression studies indicated that LP cells are in their gene expression pattern much more similar to HRS cells and the lymphoma cells of T cell–rich B cell lymphoma than they are to follicular lymphomas.15 Interestingly, LP cells show indication for strong constitutive NF-κB activity, which in its extend is similar to the NF-κB activity typical for HRS cells.15 This deregulated NF-κB activity may represent an important pathogenetic mechanism in NLPHL. Thus, although there are clear differences between classical HL and NLPHL in their gene expression, the lymphoma cells of the two types of HL also share key features.
The Role of EBV in Hodgkin Lymphoma
HRS cells in about 40% of classical HL are latently infected by the Epstein-Barr virus (EBV). As EBV can immortalize B cells in vitro and, as all HRS cells in EBV+ cases carry the virus, indicating EBV infection as an early, primary event in the development of this disease, EBV likely plays a pathogenetic role in classical HL. Although only some of the latent EBV genes are expressed in HRS cells, these may be very important. In addition to EBV nuclear antigen 1 (EBNA1), which is essential for replication of the viral genome in proliferating cells, two latent membrane proteins —LMP1 and LMP2a—are expressed. Interestingly, LMP1 mimics an active CD40 receptor and LMP2a mimics a BCR.16,17 Thus, EBV provides two signals that are essential for the survival of GC B cells, the presumed normal counterpart of HRS cells. The specific role of EBV in the pathogenesis of EBV+ classical HL is further substantiated by the finding that essentially all cases with destructive IgV gene mutations that prevent expression of a BCR at all are EBV+.18 This indicates that GC B cells that acquired BCR-destructive mutations can survive and in rare cases give rise to HRS cells only if the cells are EBV infected, so that the BCR– B cells can be rescued from apoptosis by LMP2a expression. It has indeed been shown that EBV can rescue B cells with BCR-destructive mutations in vitro, and that LMP2a has a key role in this process.17,19,20
Multiple Mechanisms Causing Constitutive NF-κB Activity
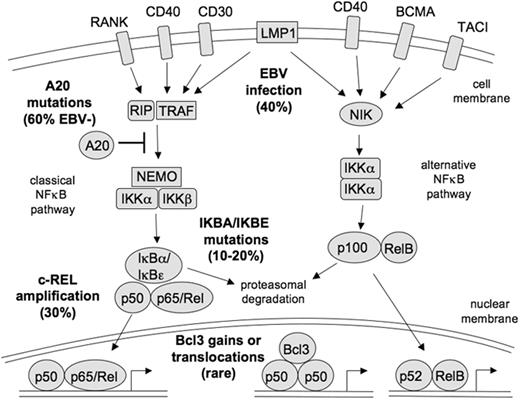
NF-κB is a transcription factor family consisting of five members, Rel, RelA (p65), RelB, p50, and p52, which can function as homo- and/or heterodimers. In the classical NF-κB pathway, the p50/p65 dimer is kept in the cytoplasm by binding to NF-κB inhibitors, in particular IκBα. Upon activation of the pathway, IKK kinases phosphorylate IκBα and thereby induce its proteasomal degradation, so that the NF-κB dimers can translocate into the nucleus and activate multiple target genes (Figure 1 ).
HRS cells consistently show strong constitutive activity of the transcription factor NF-κB,21 which is normally only transiently activated in B cells. This activation may partially be due to receptor-mediated signaling. For example, HRS cells express the TNF receptor family member CD40, which upon activation can cause NF-κB activation, and CD40 ligand-expressing T cells are found in the HL microenvironment.22 Other receptors expressed by HRS cells for which there is indication that they also are activated and cause NF-κB activation include CD30, TACI, BCMA, RANK and Notch1 (Figure 1 ).13,23,24 However, these signaling pathways are apparently not sufficient to lead to the strong constitutive NF-κB activation seen in HRS cells, because several genetic lesion have been identified in HRS cells that affect the NF-κB pathway (Figure 1 ). About 40% to 50% of classical HL cases show genomic gains or amplification of the REL gene, coding for one of the NF-κB factors.25,26NFKBIA, encoding for IκBα, is inactivated by somatic mutations in about 15% to 20% of classical HL, and another member of the IκB family, IκBε, is also mutated in a few cases.27–31 In a search for additional regulators of NF-κB activity that might be affected by genetic lesions, we and others recently identified TNFAIP3 as a novel tumor suppressor gene in HL pathogenesis that is frequently inactivated by somatic mutations.32,33 A20, the protein encoded by the TNFAIP3 gene, has ubiquitinase and deubiquitinase functions and acts upstream of the IKK kinases as a negative regulator of NF-κB signaling. We identified somatic and clonal inactivating mutations in about 40% of cases,33 and Kato and coworkers found mutations in about 30% of cases.32 Importantly, there was a striking inverse correlation between presence of EBV in HRS cells and TNFAIP3 mutation. Twelve percent of the EBV+ cases analyzed showed mutations (and these were “only” missense mutations), whereas among EBV– cases TNFAIP3 mutations were found in 70% of the cases (mostly clearly destructive and affecting both alleles).33 This indicates that EBV infection and A20 inactivation represent alternative mechanisms for NF-κB activation in HRS cells, and hence also further supports a pathogenetic role of EBV in the EBV+ cases. The function of A20 as a tumor suppressor in HRS cells was further validated by functional studies, showing that reexpression of A20 in A20-deficient HL cell lines causes reduced expression of NF-κB target genes and negatively affects cell survival.32,33 Similarly, it was shown that growth of a HL cell line with reestablished A20 expression impaired growth of these cells in a mouse model.32
As our recent gene expression studies of LP cells of NLPHL had revealed strong NF-κB activity also in the LP cells, we wondered whether similar genetic lesions cause the NF-κB activity in HRS and LP cells. However, a molecular analysis of LP cells from 10 cases of NLPHL revealed that inactivating mutations in the NFKBIA and TNFAIP3 gene are very rare, if they occur at all (manuscript submitted). Nevertheless, the role of TNFAIP3 as a tumor suppressor gene goes beyond classical HL, as mutations of this gene were also found in primary mediastinal B-cell lymphoma, diffuse large B-cell lymphoma and various types of mucosa-associated lymphoid tissue (MALT) lymphoma.32–35 All these lymphomas are also characterized by constitutive NF-κB activity.
HRS Cells in Their Microenvironment
HRS cells grow in a typical microenvironment that is composed of many different types of leukocytes, including B cells, T cells, plasma cells, eosinophils and mast cells. This microenvironment is most likely essential for HRS cell survival, as indicated from the difficulty to grow HRS cells in culture or in immunodeficient mice. HRS cells appear to regulate their microenvironment and specifically attract many of the infiltrating cells by the secretion of cytokines and chemokines.3 For example, HRS cells attract TH2-type T helper cells and Treg cells by secretion of CCL5, CCL17, and CCL22, and the secretion of IL5, IL9, CCL5, CCL28 and granulocyte-macrophage colony-stimulating factor by HRS cells attracts eosinophils into the HL microenvironment.3
CD4+ T cells usually represent the largest population of cells in the lymphoma tissue. A fraction of these cells are CD4+ T helper cells, and these may play a pathogenetic role by stimulating the survival and growth of HRS cells. Indeed, although HRS cells lost most B cell–typical genes, they retained expression of MHC class II, CD40, CD80 and CD86, key molecules for an interaction of B cells with T helper cells.36,37 In support of such an interaction, HRS cells are typically in direct contact with CD40 ligand-expressing CD4+ T cells.22 However, it has recently become clear that many of the CD4+ T cells in classical HL are not helper but regulatory T cells.38,39 These cells may also have a pathogenetic role, as there is indication that they suppress cytotoxic T cells and thereby inhibit an attack of these T cells against HRS cells.38,39
Conclusions
In recent years, considerable progress has been made in our understanding of the pathogenesis of HL. Not only the global loss of the B cell phenotype of HRS cells was revealed, but several factors contributing to this dramatic “reprogramming” have been revealed and a picture is emerging of a very much deregulated transcription factor network that is the basis for this reprogramming. It remains to be determined whether this loss of the B-cell phenotype is linked to the derivation of HRS cells from crippled GC B cells and/or is a byproduct of the transforming events in these cells and is essential for HRS cell generation. Regarding the cellular origin of HRS cells, it remains to be clarified whether HRS cells in the different subgroups of classical HL differ in the specific subset or differentiation stage of the B cells from which they derive. For example, there is a pathological overlap between primary mediastinal large B-cell lymphoma and specifically classical HL of the nodular sclerosis subtype; whether both may derive from thymic B cells is being discussed.40 Several recurrent genetic lesions have been identified in HRS cells, and it is notable that most of them affect members of the JAK/STAT or NF-κB signaling pathways.3 This is a strong indication for the key pathogenetic role of the constitutive activity of these pathways for HRS cell survival and proliferation. Finally, HRS cells attract many cells into the lymphoma tissue resulting in an inflammatory microenvironment. This environment likely promotes the survival of HRS cells and helps them to escape an attack from cytotoxic T or NK cells. A better understanding of the essential cellular interactions may offer novel approaches for a targeted therapy of this malignancy.41
The NF-κB signaling pathway and its activation in HRS cells. In the classical NF-κB signaling pathway, activation of diverse receptors leads via TRAFs (TNF receptor associated factors), often in association with the receptor interacting protein (RIP), to activation of the IKK complex, which consists of IKKα, IKKβ and NEMO. The IKK complex phosphorylates the NF-κB inhibitors IκBα and IκBε. This marks them for proteasomal degradation, thereby releasing the NF-κB factors (p50/p65 or p50/Rel heterodimers) and allowing their nuclear translocation. The signal transduction can also be inhibited by A20, which removes activating ubiquitins from RIP and TRAFs and adds Lys-48–linked ubiquitins to these molecules to mark them for proteasomal degradation. In the alternative NF-κB pathway, receptor activation leads to stimulation of the kinase NIK, which then activates an IKKα complex. Activated IKKα processes p100 precursors to p52 molecules, which then translocate as p52/RelB NF-κB heterodimers into the nucleus. HRS cells show constitutive activity of both the classical and alternative NF-κB signaling pathway, which is a central mechanism in HL pathogenesis. The NF-κB activity in HRS cells is mediated by diverse mechanisms, eg, receptor signaling through CD40, RANK, BCMA, and TACI, and signaling through the EBV-encoded latent membrane protein 1 in EBV-positive cases. Genetic lesions contributing to NF-κB activity involve genomic REL amplifications, destructive mutations in the genes of the NF-κB inhibitors IκBα and IκBε, gains or translocations of Bcl-3, and inactivating mutations in the NF-κB inhibitor A20, the latter mainly in EBV-negative cases. The frequency of genetic lesions and viral infections in classical HL cases is indicated. Adapted from Küppers.3
The NF-κB signaling pathway and its activation in HRS cells. In the classical NF-κB signaling pathway, activation of diverse receptors leads via TRAFs (TNF receptor associated factors), often in association with the receptor interacting protein (RIP), to activation of the IKK complex, which consists of IKKα, IKKβ and NEMO. The IKK complex phosphorylates the NF-κB inhibitors IκBα and IκBε. This marks them for proteasomal degradation, thereby releasing the NF-κB factors (p50/p65 or p50/Rel heterodimers) and allowing their nuclear translocation. The signal transduction can also be inhibited by A20, which removes activating ubiquitins from RIP and TRAFs and adds Lys-48–linked ubiquitins to these molecules to mark them for proteasomal degradation. In the alternative NF-κB pathway, receptor activation leads to stimulation of the kinase NIK, which then activates an IKKα complex. Activated IKKα processes p100 precursors to p52 molecules, which then translocate as p52/RelB NF-κB heterodimers into the nucleus. HRS cells show constitutive activity of both the classical and alternative NF-κB signaling pathway, which is a central mechanism in HL pathogenesis. The NF-κB activity in HRS cells is mediated by diverse mechanisms, eg, receptor signaling through CD40, RANK, BCMA, and TACI, and signaling through the EBV-encoded latent membrane protein 1 in EBV-positive cases. Genetic lesions contributing to NF-κB activity involve genomic REL amplifications, destructive mutations in the genes of the NF-κB inhibitors IκBα and IκBε, gains or translocations of Bcl-3, and inactivating mutations in the NF-κB inhibitor A20, the latter mainly in EBV-negative cases. The frequency of genetic lesions and viral infections in classical HL cases is indicated. Adapted from Küppers.3
Disclosure Conflict-of-interest disclosure: The author declares no competing financial interests. Off-label drug use: None disclosed.
References
Author notes
Institute of Cell Biology (Cancer Research), University of Duisburg-Essen, Essen, Germany