Abstract
The clinical course of patients with chronic lymphocytic leukemia (CLL) is heterogeneous, with some patients experiencing rapid disease progression and others living for decades without requiring treatment. Clinical features and molecular/biologic factors such as ZAP-70, immunoglobulin heavy chain (IGHV) gene mutation status, and cytogenetic abnormalities on fluorescent in situ hybridization (FISH) have been found to be robust predictors of treatment-free survival and overall survival among newly diagnosed patients. Beyond their widely recognized value for providing insight into disease biology and utility for stratifying patient risk in clinical trials, these prognostic tools play an important role in the current counseling and management of patients with CLL. Recent studies have focused on how to combine the results of multiple prognostic assays into an integrated risk stratification system and explored how these characteristics influence response to treatment. This chapter reviews the available tools to stratify patient risk and discusses how these tools can be used in routine clinical practice to individualize patient counseling, guide the frequency of follow-up, and inform treatment selection.
Introduction: Why Are Prognostic Tools Necessary?
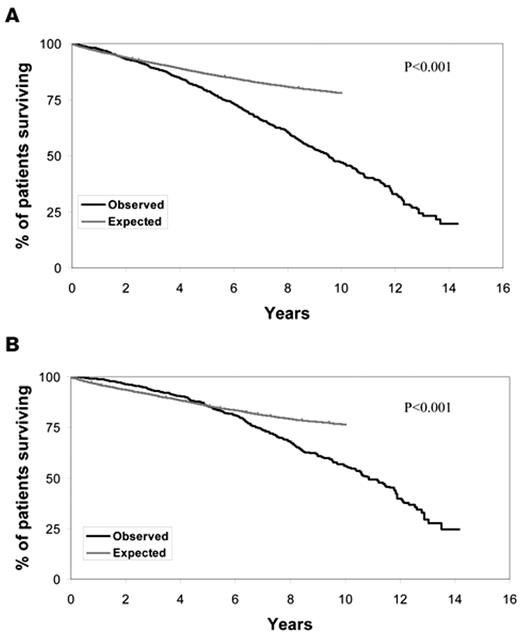
Chronic lymphocytic leukemia (CLL) is one of the most common lymphoid malignancies, accounting for approximately 11% of all hematologic neoplasms.1 Approximately 100,000 individuals in the United States are living with CLL, and the life expectancy for these patients is substantially shorter than age-matched individuals in the general population (Figure 1 ). A majority of patients with CLL are now diagnosed at the earliest stage of the disease (Rai stage 0).2,3 The clinical course of such early-stage patients is heterogeneous with some experiencing rapid disease progression and others living for decades without requiring treatment. The available evidence indicates that treatment of unselected early stage patients with alkylating agents at diagnosis offers no survival advantage over treatment at the time of disease progression.4,5 Based on this evidence, standard management of patients with newly diagnosed, early-stage CLL is to pursue a strategy of “active monitoring” that defers treatment until disease progression.
While evidence based, this approach can be profoundly difficult for patients who have been taught the mantra that early detection and treatment are the keys to improving outcome in human malignancy. The patient with CLL is given a diagnosis of leukemia and told it has been identified at the earliest stage but informed that the management strategy is to wait until it gets worse before starting treatment. This approach often leads to profound anxiety and emotional distress for patients with CLL.6 Studies suggest that, even years after diagnosis, a majority of patients with CLL think about their leukemia every day and indicate that “uncertainty about whether my CLL will progress” is among the most important sources of anxiety.7 While physicians often try to reassure patients that “CLL is the ‘good’ leukemia,” such euphemisms can imply that the oncologist views the illness as unimportant relative to other cancers and appear to increase rather than reduce patient’s distress.7 Such statements are also inaccurate as many patients with early-stage CLL experience rapid disease progression and premature death.
Prognostic tools represent an alternative approach to such generic platitudes and allow the physician to provide patients personalized counseling based on their individual risk. Thus, beyond their widely recognized value for providing insight into disease biology and utility for stratifying patient risk in clinical trials, prognostic tools play an important role in the current counseling and management of patients with CLL. These attributes of prognostic tools have culminated in robust, international efforts to study the relevant biology of the CLL B-cell and its environment, which have identified biomarkers with predictive power far beyond that of traditional clinical factors alone. In this paper we review the available tools for patient risk stratification and discuss how these tools can be used in routine clinical practice to individualize patient counseling, guide the frequency of follow-up, and inform treatment selection.
Starting with the Correct Diagnosis
In order to provide accurate prognostic information it is first essential to establish the correct diagnosis. Immunophenotyping of peripheral blood lymphocytes allows CLL to be distinguished from other low-grade lymphomas in most cases. Patients with a CD5+ lymphoma that expresses B-cell markers in a pattern atypical for CLL should undergo an appropriate work up for non-Hodgkin’s lymphoma (marginal zone, mantle cell, lymphoplasmacytic, etc) including fluorescent in situ hybridization (FISH) analysis with an IGH probe and, if possible, histopathologic tissue evaluation.8 In our experience, a diagnosis other than CLL will be found in most cases.8
Among individuals with classic CLL immunophenotype (co-expression of CD5, CD23, CD20 [dim], surface Ig [dim]), B-cell count and physical exam findings are used to distinguish between CLL, monoclonal B-cell lymphocytosis (MBL), and small lymphocytic lymphoma (SLL; Table 1 ).9,10 While some classification systems continue to distinguish between CLL and SLL, it has subsequently been recognized that these are clinically and biologically one entity11; and they were grouped as such by the WHO in 1999.12 Recent evidence suggests that the distinction between CLL and MBL has clinical implications with respect to risk of progression, justifying the clinical use of this classification system in routine practice.13,14 The topic of MBL is discussed in detail in the accompanying paper by AC Rawstron, beginning on page 430.
Predicting Outcome Using Clinical and Basic Laboratory Factors
The clinical staging systems developed by Rai and Binet were the first widely employed prognostic tools in CLL.15,16 Based simply on physical exam and a complete blood count, these tools are widely available, inexpensive, and reliable predictors of patient survival. The characteristics used to classify patient stage and the expected survival by Rai stage category in 2009 are shown in Table 2 . Despite their utility, there remains significant heterogeneity in the clinical behavior of patients within the individual stage categories. This limitation is compounded by the fact that a majority of patients are now diagnosed with Rai stage 0 disease where stage alone does not adequately predict the risk of progression for a given patient.
To address this limitation of clinical staging, the ability of other basic laboratory parameters to predict clinical outcome have been explored. The lymphocyte doubling time (LDT), calculated by determining the number of months it takes the absolute lymphocyte count (ALC) to double in number, is a marker of disease kinetics that has been found to correlate with both progression-free (PFS) and overall survival (OS).17 Even simpler than the LDT, the ALC has also been shown to predict both PFS and OS.18,19 One recent study found a median PFS of 20 months for Binet stage A patients with a LDT ≤ 12 months compared with 75 months for those with a LDT > 12 months (P < .001). The same study found a median PFS of 17 months for patients with an ALC > 30 × 109/L compared with 88 months for those with an ALC < 30 × 109/L.18
Serum beta 2 microglobulin (B2M) is another laboratory parameter that correlates with clinical outcome in patients with CLL.18,19 In one recent study the median PFS of patients with a B2M > 3.5 mg/L was 13 months compared with 75 months for those with a B2M < 3.5 mg/L (P < .001).18
In an effort to develop a simple method of risk stratification that integrates the above clinical and laboratory factors into a single model, Wierda and colleagues recently analyzed clinical outcome in a large series of patients cared for at the MD Anderson Cancer Center. This analysis identified six factors (age, stage [Rai], sex, ALC, B2M, and number of lymph node regions involved) that were independently associated with patient survival.19 These relatively easy to obtain factors were then combined in a prognostic index that was able to predict survival more accurately than clinical stage alone.19 This prognostic index was subsequently validated in two independent series of patients20,21 that further demonstrated the index remained useful when applied exclusively to Rai stage 0 patients and was able to predict time to treatment as well as OS.20
Although multiple molecular/biologic characteristics of CLL B cells have been shown to be powerful predictors of survival in patients with CLL (discussed below), many of these biologic assays are expensive, difficult to standardize, and not widely available. The prognostic index developed by Wierda and colleagues provides an advance to those patients who do not have access to such tests and incorporates most of the readily available clinical and basic laboratory factors into a single prognostic model. While useful for predicting outcome, the limitation of such clinical prognostic tools is that the factors they are based on (eg, age, sex, ALC) do not provide the insights into disease biology that are necessary to develop more targeted/ effective treatments. Nonetheless, experience from other B-cell malignancies suggests that the combination of clinical prognostic tools and relevant molecular assays will lead to more precise prediction of patient outcome than either strategy alone.22
Predicting Outcome Using Molecular/ Biologic Factors
The fact that the immunoglobulin heavy chain genes of CLL B cells had undergone somatic hypermutation in roughly one half of patients was recognized around 15 years ago.23 The prognostic importance of this characteristic was independently reported by two groups in 1999.24,25 These studies demonstrated that CLL patients with mutated immunoglobulin genes had an OS expectation greater than 20 years, while those with unmutated immunoglobulin genes had median survivals in approximately the 8-year range.24,25 The ability of IGHV mutation status to stratify OS remained when applied exclusively to early-stage patients.25 After this landmark finding, there has been an explosion in the discovery of other biologic and molecular characteristics of the CLL B cell that relate not only to disease biology but also to patient’s PFS and OS. The four molecular/biologic features that have the best track record for use as clinical prognostic parameters are IGHV mutation status,24,25 recurrent cytogenetic abnormalities as identified by FISH testing,26 zeta-associated protein (ZAP) 70 expression27,28 and CD38 protein expression.24,29 Each of these biomarkers has been shown to be a predictor of time to treatment and OS on univariate analysis.
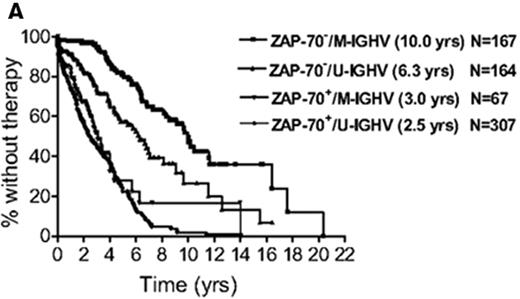
With the near simultaneous discovery of multiple prognostic factors, determining which are most useful and how best to combine the often discordant results of multiple tests has been challenging. To address this issue, the CLL Research Consortium assessed the relative value of ZAP-70, CD38, and IGHV mutation status for predicting time to treatment in a study of over 1000 CLL patients.28 The results suggest patients can be categorized into low-, intermediate-, and high-risk groups based on ZAP-70 and IGHV mutation status where ZAP-70–positive patients are high risk regardless of IGHV mutation status and patients who are ZAP-70 negative can be classified as low (IGHV mutated) or intermediate (IGHV unmutated) risk based on IGHV status (Figure 2A ).28 CD38 had no incremental prognostic value for predicting time to treatment independent of ZAP-70 and IGHV analysis.28 Despite the confirmed ability of ZAP-70 to predict patient outcome in numerous studies, variation in the anti–ZAP-70 antibody used in the assay, the staining procedure, and the gating strategy to analyze results have made standardization and clinical application of this test difficult. International standardization of the ZAP-70 assay remains necessary before widespread clinical use can be recommended outside of academic research laboratories.30
Cytogenetic abnormalities as assessed by FISH have also been a widely applied prognostic tool since Döhner and colleagues reported a hierarchical classification system to predict clinical outcome using FISH.26 In their analysis, patients with del(17p13) had a median survival from diagnosis of 2 to 3 years, as compared to 6 to 7 years for those with del(11q22), about 9 years for those with trisomy 12 or normal karyotype, and about 11 years for those with del 13q14 as a sole abnormality.26 Subsequent prospective studies confirmed these results but also demonstrated that new cytogenetic abnormalities (ie, karyotype evolution) are acquired during follow-up in more than 25% of patients over a 5-year interval.31 Karyotype evolution was associated with short survival; however, this difference in survival was limited to patients who acquired a del(17p13) or del(11q22) in whom median survival after karyotype evolution was only 1.3 years.32
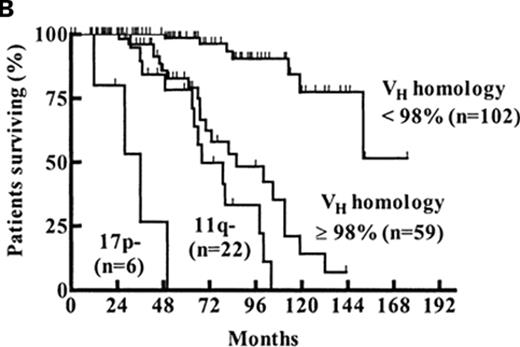
After their seminal initial report on the prognostic value of FISH in CLL,26 the group from Ulm subsequently evaluated the relative value of FISH, CD38, and IGHV mutation status in a study of 189 Binet stage A patients.33 The results suggested Binet A patients could be categorized into low-, intermediate-, and high-risk groups based on the combination of FISH and IGHV mutation status where patients with del(17p13) were high risk regardless of IGHV mutation status, patients with del(11q22) and/or unmutated IGHV genes were intermediate risk, and patients with mutated IGHV in the absence of del(17p13) or del(11q22) were low risk (Figure 2B ).33
Follow-up studies have provided further insight into the biology and clinical behavior of patients with del(17p13). In a recent report from the MD Anderson and Mayo Clinic evaluating the natural history of 99 consecutive, chemotherapy-naïve patients with del(17p13), significant heterogeneity in clinical outcome was observed.34 Among asymptomatic del(17p13) patients with Rai stage 0-II disease, a shorter time to treatment was observed among those with Rai stage ≥ 1 or unmutated IGHV mutation status. The 3-year treatment-free survival rates among patients with 0, 1, or 2 of these factors were 100%, 54%, and 0%, respectively (P < .001).34 Similarly, shorter OS was observed among del(17p13) patients who had Rai stage ≥ 1, unmutated IGHV mutation status, or ≥ 25% of nuclei affected by del(17p13). The 3-year OS rates among patients with 0–1, 2, or 3 of these risk factors were 95%, 74%, and 22%, respectively (P < .001).34 Several studies recently published or presented in abstract form indicate that mutation of TP53 is a prognostic parameter independent of del(17p13)35–37 with some36 but not all35 suggesting that the clinical implications of del(17p13) on FISH may be influenced by whether or not the TP53 gene on the remaining allele is functional. Thus, in one study of 529 patients, the 3-year survival of those with bi-allelic insufficiency of TP53, mono-allelic insufficiency of TP53 (either by TP53 mutation or deletion), and no abnormalities of TP53 were 23%, 52% and 80%, respectively (P < .00001).36
In addition to IGHV, FISH, ZAP-70, and CD38, an extensive number of other prognostic parameters have been identified including overexpression of the CLLU1 gene, CD49d protein expression, telomere length/telomerase activity, expression of Bcl-2 family member proteins, markers of angiogenesis, soluble CD23, Rel A DNA binding, thymi-dine kinase, lymphocyte smudge cell percentage, genomic complexity, and lipoprotein lipase gene expression (reviewed in van Bockstaele et al38). While these assays provide important insight into disease biology and in some cases may represent potential therapeutic targets, they are not routinely available or widely used for clinical prognostication at the present time.
Response to Treatment as a Prognostic Marker
Patient response to treatment has long been recognized as a predictor of survival in most human malignancies. Numerous studies in CLL have demonstrated that individuals with a better response to treatment (complete remission vs partial remission or minimal residual disease [MRD]-negative remission vs MRD-positive remission) have a longer response duration and OS. While there is no doubt response to treatment identifies patients with a better outcome once treatment is required, this metric obviously has no utility in newly diagnosed, early-stage patients since they are observed rather than treated. Even for patients requiring therapy, response to treatment cannot be assessed until therapy has been completed and, as such, it is a retrospective prognostic parameter that is not useful until after patients have been exposed to toxic therapy. While some have interpreted the relationship between response to treatment and survival in CLL patients to indicate that more aggressive first-line treatment or consolidation therapy for patients with residual disease will improve outcome, this is a hypothesis that must be tested in randomized trials.
Use of Prognostic Markers to Predict Response to Treatment
In addition to their ability to stratify treatment-free survival and OS, some biomarkers may also be useful for predicting response (or resistance) to specific therapeutic agents. The presence of del(17p13) and/or abnormal TP53 function have consistently been shown to identify CLL patients who are unlikely to respond to purine nucleoside analogues (fludarabine, pentostatin, cladribine), alkylating agents (chlorambucil, cyclophosphamide, bendamustine), and combinations of these drug classes with or without rituximab.39,40 In contrast, individuals with TP53 abnormalities do appear to respond to other therapies such as high-dose methylprednisolone, alemtuzumab, flavopiridol, or allogeneic transplant although, outside of allogeneic transplant, the responses are generally short lived.41–44 Although the weight of the evidence does not suggest IGHV mutation status predicts whether or not patients will achieve a complete remission in response to treatment, significant evidence suggests mutation status is a powerful predictor of the duration of response.39,40,45,46 The ability of ZAP-70 and CD38 to predict response to treatment and PFS/ OS after treatment is less clear (Table 3 ). A better understanding of how specific molecular features relate to leukemia cell biology and additional studies of how such characteristics influence response to treatment may ultimately allow physicians to individualize treatment selection for CLL patients in a manner that maximizes the potential for benefit and reduces the likelihood of unnecessary toxicity.
How to Use Prognostic Information in Clinical Practice
Determining clinical stage remains the first step in predicting outcome for patients with newly diagnosed CLL. For patients with Rai stage 0 or Binet stage A disease, risk can be further sub-divided using additional prognostic tools. At present, there are at least four ways such prognostic tools should be used to complement clinical staging in routine clinical practice. First, they should be employed to subdivide patients with Rai stage 0 or Binet stage A as having low-, intermediate-, or high-risk disease (Rai 0-low risk; Rai 0-intermediate risk; Rai 0-high risk). While using a combination of molecular biomarkers as shown in Figure 2A or 2B appears to be the most accurate method for such classification, the prognostic index developed by Wierda and colleagues19 or lymphocyte doubling time17,18 represent alternative strategies for patients who do not have access to these assays. Once stratified, patients should be provided individualized counseling regarding typical anticipated treatment-free and OS. Patients with Rai 0-low-risk disease can be reassured while a more cautious tenor should be taken in the counseling of Rai 0-intermediate and Rai 0-high risk patients. Second, the frequency of follow-up visits for disease monitoring can be individualized based on patient risk. We currently recommend individuals with Rai 0-high-risk disease, such as those with del(17p13), be seen every 3 to 6 months. Individuals with Rai 0-intermediate risk can be seen every 6 to 12 months, while those with Rai-0 low-risk can be seen on an annual basis. Third, although no patient with early-stage, asymptomatic disease should be routinely treated based on the results of prognostic testing, these tools should be used to identify early-stage patients with high-risk disease who may be candidates for treatment in well-designed clinical trials. Fourth, the presence of del(17p13) on FISH testing can be used to help guide therapy selection once patients meet the conventional indications for treatment. Given their inferior response to purine analogues, alkylating agents, and purine/alkylator combinations, participation in well-designed clinical trials designed specifically for patients with del(17p13) should be the preferred approach for CLL patients with this cytogenetic defect. Outside of trials, patients with del(17p13) appear to respond to high-dose methylpredniso-lone and alemtuzumab based strategies although responses are often short lived. Appropriate younger patients with del(17p13) are candidates for allogeneic stem cell transplant in either first or second remission.44,47 In this regard, it is reasonable to initiate a donor search at the time first-line therapy is initiated for younger patients with del(17p13) who will be candidates for this strategy given their high risk of non-response to first-line therapy and/or the frequently short duration of response.
Distinguishing between monoclonal B-cell lymphocytosis (MBL), chronic lymphocytic lymphoma (CLL), and small lymphocytic lymphoma (SLL).
| . | Clonal B-cells of CLL phenotype . | Peripheral blood B-cell count <5 × 109/L . | Lymphadenopathy or hepatosplenomegaly . |
|---|---|---|---|
| Reprinted with permission from Shanafelt T, Hanson CA. Monoclonal B-cell lymphocytosis: definitions and natural history. Leuk Lymphoma. 2009;50:493–497.48 | |||
| MBL | + | + | − |
| SLL | + | + | + |
| CLL | + | − | +/− |
| . | Clonal B-cells of CLL phenotype . | Peripheral blood B-cell count <5 × 109/L . | Lymphadenopathy or hepatosplenomegaly . |
|---|---|---|---|
| Reprinted with permission from Shanafelt T, Hanson CA. Monoclonal B-cell lymphocytosis: definitions and natural history. Leuk Lymphoma. 2009;50:493–497.48 | |||
| MBL | + | + | − |
| SLL | + | + | + |
| CLL | + | − | +/− |
Median survival by Rai stage.
| Rai Stage . | Characteristic . | Median survival: Original Report (n = 125)15 . | Median survival: Mayo Clinic CLL Database* (n = 2397) . |
|---|---|---|---|
| *All individuals with CLL seen in the Mayo Clinic Division of Hematology since 1995. | |||
| **Hg < 11 g/dL | |||
| †Platelet count <100 × 109/L | |||
| 0 | Lymphocytosis only | 150 months | 130 months |
| I | Lymphadenopathy | 101 | 106 |
| II | Organomegaly | 71 | 88 |
| III | Anemia** | 19 | 58 |
| IV | Thrombocytopenia† | 19 | 69 |
| Rai Stage . | Characteristic . | Median survival: Original Report (n = 125)15 . | Median survival: Mayo Clinic CLL Database* (n = 2397) . |
|---|---|---|---|
| *All individuals with CLL seen in the Mayo Clinic Division of Hematology since 1995. | |||
| **Hg < 11 g/dL | |||
| †Platelet count <100 × 109/L | |||
| 0 | Lymphocytosis only | 150 months | 130 months |
| I | Lymphadenopathy | 101 | 106 |
| II | Organomegaly | 71 | 88 |
| III | Anemia** | 19 | 58 |
| IV | Thrombocytopenia† | 19 | 69 |
Current interpretation of data regarding the ability of biomarkers to predict response to purine nucleoside analogues (PNA) and alkylating agents ± rituximab.
| Prognostic factor . | Predicts poor response to PNA & alkylating agents ± rituximab . | Predicts shorter PFS/OS after treatment with PNA & alkylating agents ± rituximab . | Selected References . |
|---|---|---|---|
| del(17p13) or TP53 defects | Yes | Yes | Marker as predictor of response: |
| Support: 36,39,40,49–52 | |||
| Against: 45,53 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 36,39,40,45,50–53 | |||
| del(11q22) | Yes/No | Yes | Marker as predictor of response: |
| Support: 39 | |||
| Against: 45,50,52,53 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 39,45,53 | |||
| Against: 50,52 | |||
| Unmutated IGHV | No | Yes | Marker as predictor of response: |
| Against: 46,50,53,54 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 39,40,45,46,52,54,55 | |||
| Against: 50,53 | |||
| ZAP-70+ | No | Yes/No | Marker as predictor of response: |
| Support: 52 | |||
| Against: 50,53,54 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 52,55 | |||
| Against: 50,53,54 | |||
| CD38+ | No | No | Marker as predictor of response: |
| Against 50,53,54 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 55 | |||
| Against: 50,52–54 |
| Prognostic factor . | Predicts poor response to PNA & alkylating agents ± rituximab . | Predicts shorter PFS/OS after treatment with PNA & alkylating agents ± rituximab . | Selected References . |
|---|---|---|---|
| del(17p13) or TP53 defects | Yes | Yes | Marker as predictor of response: |
| Support: 36,39,40,49–52 | |||
| Against: 45,53 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 36,39,40,45,50–53 | |||
| del(11q22) | Yes/No | Yes | Marker as predictor of response: |
| Support: 39 | |||
| Against: 45,50,52,53 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 39,45,53 | |||
| Against: 50,52 | |||
| Unmutated IGHV | No | Yes | Marker as predictor of response: |
| Against: 46,50,53,54 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 39,40,45,46,52,54,55 | |||
| Against: 50,53 | |||
| ZAP-70+ | No | Yes/No | Marker as predictor of response: |
| Support: 52 | |||
| Against: 50,53,54 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 52,55 | |||
| Against: 50,53,54 | |||
| CD38+ | No | No | Marker as predictor of response: |
| Against 50,53,54 | |||
| Marker as predictor of PFS/OS: | |||
| Support: 55 | |||
| Against: 50,52–54 |
Overall survival of CLL patients relative to age-matched controls. A. Figure shows the survival from diagnosis of 2474 Mayo Clinic patients diagnosed with CLL since January 1995 as compared with the age-matched general Minnesota population. P < .001. B. Figure shows the survival from diagnosis of 1282 Mayo Clinic patients diagnosed with Rai stage 0 CLL since January 1995 as compared with the age matched general Minnesota population. P < .001.
Overall survival of CLL patients relative to age-matched controls. A. Figure shows the survival from diagnosis of 2474 Mayo Clinic patients diagnosed with CLL since January 1995 as compared with the age-matched general Minnesota population. P < .001. B. Figure shows the survival from diagnosis of 1282 Mayo Clinic patients diagnosed with Rai stage 0 CLL since January 1995 as compared with the age matched general Minnesota population. P < .001.
Combining biomarkers to predict patient outcome. A: ZAP-70 and IGHV Mutation Status.28 Figure shows time from diagnosis to first treatment based on the combination of ZAP-70 and IGHV mutation status. Reprinted with permission of the American Society of Hematology. This research was originally published in Blood by Rassenti et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112:1923–1930 © the American Society of Hematology. B: FISH and IGHV Mutation Status.33 Figure shows survival from diagnosis among Binet stage A patients based on combination of FISH and IGHV mutation status. Patients with del(17p13) or del(11q22) are classified by FISH category regardless of the results of IGHV testing. Patients without del(17p13) or del(11q22) are stratified as unmutated (VH homology ≥ 98%) or mutated (VH homology<98%) based on IGHV testing. Reprinted with permission of the American Society of Hematology. This research was originally published in Blood by Krober et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416 © the American Society of Hematology.
Combining biomarkers to predict patient outcome. A: ZAP-70 and IGHV Mutation Status.28 Figure shows time from diagnosis to first treatment based on the combination of ZAP-70 and IGHV mutation status. Reprinted with permission of the American Society of Hematology. This research was originally published in Blood by Rassenti et al. Relative value of ZAP-70, CD38, and immunoglobulin mutation status in predicting aggressive disease in chronic lymphocytic leukemia. Blood. 2008;112:1923–1930 © the American Society of Hematology. B: FISH and IGHV Mutation Status.33 Figure shows survival from diagnosis among Binet stage A patients based on combination of FISH and IGHV mutation status. Patients with del(17p13) or del(11q22) are classified by FISH category regardless of the results of IGHV testing. Patients without del(17p13) or del(11q22) are stratified as unmutated (VH homology ≥ 98%) or mutated (VH homology<98%) based on IGHV testing. Reprinted with permission of the American Society of Hematology. This research was originally published in Blood by Krober et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416 © the American Society of Hematology.
Disclosures Conflict-of-interest disclosure: The author receives research funding from Genentech, Hospira, Polyphenon E International, Celgene, Cephalon, Bayer Health Care Pharmaceuticals, and also serves on an advisory committee/ board of directors for Hospira. Off-label drug use: None disclosed.
Acknowledgments
References
Author notes
Department of Internal Medicine, Division of Hematology, Mayo Clinic, Rochester, MN