Abstract
The last decade has witnessed an abundance of information detailing the genetic diversity of the RH locus which has exceeded all estimates predicted by serology. Well over 120 RHD and over 60 different RHCE alleles have been documented, and new alleles are still being discovered. For clinical transfusion medicine, RH genetic testing can now be used to determine RHD zygosity, resolve D antigen status, and detect altered RHD and RHCE genes in individuals at risk for producing antibodies to high-incidence Rh antigens, particularly patients with sickle cell disease (SCD).
Transfusion of patients with sickle cell disease (SCD) represents a significant challenge in clinical transfusion medicine with red blood cell (RBC) alloimmunization a primary and serious complication of transfusions. A major cause of alloimmunization in patients with SCD is the disparate distribution of red cell antigens between donors, who are primarily of European ancestry, and patients with SCD, who are primarily of African ancestry. Management of alloimmunization in SCD has been the subject of much debate,1,2 and currently there is no standard approach. Over two thirds of alloantibodies formed by patients with SCD have Rh blood group specificities. Many programs transfuse patients with SCD with RBCs that are phenotype-matched for D, C/c, E/e, and K, and some also supply RBCs from African-American donors when possible. Although these approaches reduce the incidence of alloantibody production, patients still become alloimmunized. Genetic analysis of patients who develop antibodies in the face of conventional antigen matching has revealed that these patients carry altered RH alleles.
RH Genes and Rh Proteins
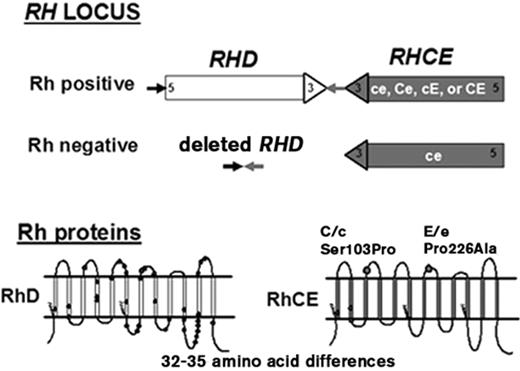
Two genes, RHD and RHCE, lie in close proximity on chromosome 1 and encode 416–amino acid Rh proteins: one encodes the D antigen, and the other encodes CE antigens in various combinations (ce, cE, Ce, or CE) (Figure 1 ). Each gene has ten exons; they are 97% identical and encode proteins that differ by 32 to 35 amino acids (Figure 1 ). This contrasts with most blood group antigens, which are encoded by single genes with alleles that differ by only one or a few amino acids. For comprehensive reviews, see Westhoff3 and Avent.4
The conventional RH genes shown in Figure 1 are found in all population groups, although with different frequencies. Commercial antibody reagents detect expression of the five principal Rh antigens: D, C, c, E, and e. Variant RHD and RHCE alleles encode Rh proteins with amino acid changes that cannot be distinguished serologically, but can be recognized by the immune system as foreign. Of relevance for transfusion in patients with SCD, RH alleles encoding altered C and e antigens are frequent in African ethnic groups.5,6 Some patients with SCD are at risk for production of antibodies to both the RhCE and RhD proteins, a serious and potentially life-threatening complication.
Variations of RhD
D Negative
D- (Rh-negative) phenotypes are the result of absence of the RhD protein in the erythrocyte. Exposure to RhD in a D-individual often results in production of anti-D. In Caucasians, D- results primarily from deletion of the entire RHD gene (Figure 1 ).7 In D- African Blacks, 66% have RHD that contains a 37 base pair insertion that results in a premature stop codon, 15% carry a hybrid RHD-CE-D characterized by expression of C antigen but no D, and 19% have a RHD gene deletion.8 This hybrid RHD-CE-D results from RHCE exons 3 through 7 replacing the region of RHD encoding D epitopes. The RBCs with this hybrid protein type as D- and C+.
D Positive
Most individuals who are D+ have identical RHD sequences, but approximately 2% have alleles that encode proteins with amino acid changes or have hybrid alleles encoding proteins with part of RhD fused to RhCE, or vice versa. These altered or hybrid proteins cause reduced expression of antigen, termed weak D,9 and/or can result in altered D surface epitopes, termed partial D.3,4 Some of these patients make anti-D when stimulated by conventional D+ RBCs. Although 1% to 2% of Europeans carry RHD alleles that encode weak or partial D, the frequency in African Black ethnic groups appears to be much higher but is not known.
Variations of RhCE
RHCE encodes both C/c and E/e antigens on a single protein. C and c antigens differ by four amino acids and only the amino acid change Ser103Pro is extracellular (Figure 1 ). The E and e antigens differ by one amino acid, Pro226Ala, located on the fourth extracellular loop of the protein. Mutations in RHCE result in quantitative and qualitative changes in C/c or E/e antigen expression, with altered C and e encountered most frequently. In Caucasians, altered C is associated with amino acid changes located on the first extracellular loop of RhCe and the expression of CW (Gln41Arg) or CX (Ala36Thr) antigens.3,4 Altered C is also associated with mutations that result in expression of the novel antigens JAHK (Ser122Leu)10 and JAL (Arg114Trp).11 The red cells type as C+, but patients can make apparent anti-C or anti-Ce (rhi) when stimulated.
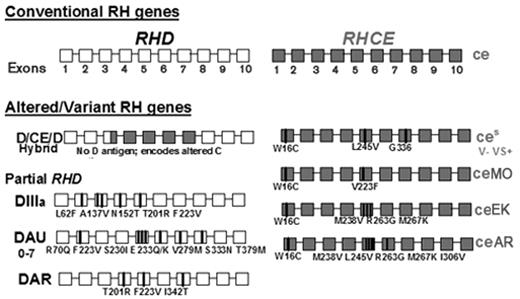
In individuals of African ancestry, altered C expression most often results from the inheritance of the RHD-CE(3–7)-D hybrid gene discussed above. This gene (Figure 2 ) does not encode D antigen, but encodes C antigen on an Rh protein background that differs from normal. The gene has an incidence of approximately 22% in African-Americans.8 It is often inherited with an RHCE allele designated ceS (Figure 2 ), which encodes altered e antigen and a V-VS+ phenotype.5 The hybrid RHD-CE(3–7)-D linked to RHCE*ceS is referred to as the (C)ceS or r′S haplotype. Red cells often type strongly C+ with monoclonal reagents and the presence of the altered C goes undetected. These patients not infrequently make alloantibodies with C-like and e-like specificities that can appear to be autoantibodies. Their red cells typically also lack the high-prevalence hrB antigen. Anti-hrB can be clinically significant, and finding compatible blood can be very difficult. Not all red cells designated hrB− by serologic testing are compatible with antibodies produced by patients with differing RH backgrounds.12,13
Altered or variant e antigen expression is associated with many other RHce alleles that have multiple mutations13,14 that are found primarily in African ethnic groups. The red cells type as e+, but individuals homozygous for these alleles make alloantibodies with e-like specificities that can appear to be autoantibodies. The red cells also lack the high-prevalence hrS antigen. Anti-hrS is a clinically significant antibody and has caused transfusion fatalities.14
As an additional complication, altered or variant RHCE*ce is often inherited with partial RHD, and examples include DIII, DAU, and DAR (Figure 2 ). The inheritance of partial D alleles with altered or variant RHCE*ce in patients with SCD puts them at risk for production of antibodies to both RhCE and RhD proteins, a potentially serious complication. A directory of RHD alleles is maintained and updated on the RhesusBase website15 and RHCE alleles are found on the NCBI human blood group mutation website.16
Clinical Considerations for Patients with SCD
The alloimmunization rate in SCD ranges from 8% to 47%, depending on the patient’s age, number of donor units transfused, and extent of phenotype matching.17–19 Transfusion is further complicated by the presence of multiple alloantibodies in as many as 45% of patients.20 The American Rare Donor Program (ARDP) has been used to search for RBCs to meet transfusion needs of patients with SCD with multiple antibodies. Between January 2005 and June 2006, approximately 33% of all the requests to the ARDP for RBCs were for alloimmunized patients with SCD.21 Alloimmunization can have severe clinical consequences, not only because there can be significant delays in finding compatible blood, but also because alloimmunization is associated with delayed hemolytic transfusion reactions (DHTR), autoantibody formation, and hyperhemolysis syndrome.
DHTRs are more common in patients with SCD compared with the general population. While DHTRs are generally mild in patients who do not have SCD, in those who have SCD they may be life-threatening, with hemoglobin levels dropping below pretransfusion levels due to bystander hemolysis of the patients’ own red cells, referred to as hyperhemolysis.22 Vichinsky and Talano report that DHTRs occurred in 11% of patients with SCD and caused profound anemia with hemolysis of both donor and autologous RBCs.19,23 In addition to life-threatening anemia, these episodes were associated with pain, acute chest syndrome and/or acute renal failure. In approximately half the cases, the direct antiglobulin test (DAT) remained negative with no detectable antibodies, making the diagnosis and management difficult.19,23 DHTRs are likely under-recognized in patients with SCD, as symptoms mimic a painful episode and supporting laboratory tests (lactate dehydrogenase, unconjugated bilirubin levels) used to diagnose DHTR are already abnormal in patients with SCD with chronic hemolysis. Fatalities from renal failure, disseminated intravascular coagulation, and stroke are occasionally reported in association with DHTRs.
Alloantibodies complicate pregnancy in patients with SCD. Antibodies in the Rh system are not often associated with severe fatal hemolytic disease of the fetus and newborn in these patients, but are associated with mild to moderate fetal anemia. At a minimum, costly monitoring of the pregnancy is needed.24,25
Antigen-matching to Reduce Alloimmunization in Patients with SCD
Over two thirds of alloantibodies formed by patients with SCD have Rh blood group specificities. Therefore, in an effort to reduce alloimmunization, some SCD programs transfuse RBCs that are antigen-matched for Rh D, C/c, and E/e and also for Kell (K) antigens.26,27 Less often, Duffy (Fya/b) and Kidd (Jka/b) antigens are also matched. Although randomized controlled trials have not been performed, extended red cell antigen–matching has been shown in numerous single institutional and prospective multicenter experiences to significantly reduce the incidence of alloantibody production in SCD.28–30 Data suggest a 40% to 90% reduction, depending on the extent of antigen matching (4 to 15 antigens), accompanied by decreased DHTRs compared with historical rates in patients with SCD. Despite this success, some institutions perform phenotype matching only after the patient develops the first alloantibody. Some argue that the data supporting reduction in alloimmunization and DHTRs are insufficient to offset the cost of the labor and resources required to perform extended phenotype matching for all cases.17 Donor availability is a major issue.
Compatible blood for patients with SCD is most likely to be found among African-American donors, whose RBCs more commonly lack C, E, K, S, Fya, Fyb, and Jkb antigens, than from Caucasian donors.31 In addition to antigen-matching for D, C/c, E/e, and K antigens, a program to direct blood from African-American donors to children with SCD was initiated in 1997 as a cooperative effort between Children’s Hospital of Philadelphia (CHOP), the Penn-Jersey American Red Cross, and the local Chapter of the Sickle Cell Disease Association of America.26 Blood donors voluntarily self-identify as African-American and agree to have their blood specifically support SCD children by selecting a special tag to attach to their donation. These donors support more than 50 chronically transfused patients with SCD at CHOP and 35 at St. Christopher’s Hospital for Children, and also support episodic needs of over 1700 patients with SCD in the community. In the decade-long experience of this program, alloimmunization rates have dramatically been reduced, but not eliminated.
RH Genotyping
DNA molecular testing methods that use polymerase chain reaction (PCR) technology were introduced a decade ago after the cloning of the genes made genetic testing for blood groups possible. Most blood group antigens are encoded by single nucleotide polymorphisms (SNPs), and assays that target these allelic polymorphisms are reproducible and highly correlated with RBC phenotype.32,33 ABO and RH are more complex. The detection of numerous silencing mutations is required for accurate typing; several regions of the genes must be sampled to detect multiple alleles, and new alleles are still being identified.
High-throughput genotyping applied to blood group typing has the potential to be the next major technological revolution in transfusion medicine. This approach offers significant cost savings in both labor and reagents compared with antigen typing by serologic methods, and expands testing to detect genetic variation of antigen expression. When coupled with recipient testing, genotyping would enable electronic selection of units antigen-matched at multiple blood group loci to significantly reduce alloimmunization. The New York Blood Center successfully collaborated with BioArray Solutions, Ltd to develop a genotyping platform for human red cell antigens. This automated platform genotypes for 32 antigens in 12 different blood group systems, all encoded by simple SNPs.34,35 The complexities and numerous alleles in the Rh blood group system have precluded testing for RH other than for the common non-variant forms of C/c and E/e antigens but automated testing is being developed. An automated test platform developed in Europe (BLOODChip, Progenika Biopharma S.A.) is now available in the United States; in addition to minor blood group antigens it includes an extensive number of variant RHD alleles.36 SNPs associated with variant expression of C/c and E/e antigen are also under development by this manufacturer (personal communication).
Currently, extensive RH genotyping is costly and time consuming and is limited to finding compatible donors in the American Rare Donor Program (ARDP) for patients with antibodies to high-prevalence Rh antigens.16 The future availability of high-throughput RH genotyping platforms will enable patients who are homozygous for altered or variant RH alleles prevalent in patients with SCD to be recognized, and compatible or genotype-matched donors to be identified.
RH Gene Polymorphisms in Patients with SCD
The Antibodies
American Red Cross molecular testing laboratory has genotyped patients with SCD from Philadelphia area hospitals, as well as samples referred from across the United States. The antibody specificity and the RH alleles found in 46 alloimmunized patients with SCD are shown in Table 1 . Group A consists of 20 patients with SCD who have become alloimmunized in our program, despite having received donor units primarily from minority donors and antigen (phenotype) matched for Rh and K. Group B consists of 26 patients with SCD who are not part of antigen-matching programs receiving random donor units. As would be expected, the patients receiving random donor units (in Group B) and not part of an antigen-matching program have a larger number of antibody specificities (108 versus 31 for an average of 4.15 antibodies/patient compared to 1.55). What is shared or consistent between the two groups is the number of complex RH antibody specificities or “apparent autoantibodies.” The antibodies identified include anti-D, -C, -e, and/or antibodies to high-prevalence antigens in patients who are serologically D+, C+ or e+, indicating that the “antigen-matched” patients in Group A who made Rh alloantibodies were not truly Rh matched.
RH Alleles
RH gene molecular analysis revealed that 23 of these 46 patients (50%) had a RHD-CE-D hybrid gene encoding altered C antigen (Table 1 and Figure 2 ). Patients with a C antigen encoded by the hybrid RHD-CE-D gene can produce alloanti-C following transfusion with conventional C+ RBCs. Of note, the D-CE-D hybrid and anti-C was found in only 3 of 20 patients who had received transfusion support primarily from African-American donors, compared with 20 of 26 alloimmunized patients with random donors. This indicates that there is benefit to providing transfusion support from African-American donors for patients with SCD who inherit this allele.
The D-CE-D hybrid discussed above is linked to an RHce allele carrying three nucleotide mutations encoding amino acid changes Trp16Cys, Leu245Val, and Gly336Cys. This allele, designated ceS, encodes an altered e antigen (Figure 2 ).5 The ceS allele was present in all 23 samples with D-CE-D (Table 1 ). Inheritance of altered C, encoded by D-CE-D, linked to altered e, encoded by ceS (referred to as the (C)ceS haplotype) is associated with production of alloantibodies with both C- and e-like specificities, termed -hrB.
Lastly, many patients produced anti-e or -hrS, although their RBCs type e+. These included 8 of 20 patients transfused with African-American donors, and 6 of 26 receiving random donors. These patients have RHce alleles with a single amino acid change (Trp16Cys)37 or multiple mutations reported primarily in African ethnic groups and designated ceMO, ceAR, ceEK, ceBl14 (Figure 2 ). RBCs from patients homozygous for any of these alleles lack the high incidence hrS antigen (hrS−). Anti-hrS is a clinically significant antibody that has caused transfusion fatalities.14 Transfusion and selection of donor units for e+ patients with anti-e are difficult to manage because e- donor units are E+, and these patients often make anti-E.
In addition to antibodies that recognize RhCe or Rhce proteins, many transfused patients with SCD also produced antibodies to RhD. These included 11 patients transfused with blood from African-American donors and 4 in the random donor group. Because anti-D can often be masked in the presence of anti-hrB, the incidence of anti-D in this group may be underestimated. Not all samples with apparent anti-D had altered or partial RHD. This may suggest that variant Rhce proteins may alter the Rh-complex in the membrane and remains under investigation. The presence of a partial RHD (DIII, DIV, DAU, or DAR) in many samples supports our observation that altered or variant RHCE*ce are often linked to alleles encoding partial D antigen.
Twenty-six patients were homozygous for mutations in RHce and had produced anti-e, and some also had mutations in RHD and had produced anti-D. These results suggest that inheritance of the RHD-CE-D gene or altered RHce with or without altered RHD, underlies Rh alloimmunization in SCD. The altered Rh proteins are not distinguished with current serologic typing reagents. Therefore, these patients are not truly Rh antigen matched. RH genotyping offers a potential solution to prevent alloimmunization by identifying patients with SCD who are homozygous for variant alleles and at risk for production of alloantibodies to Rh antigens. Donor units with the same RH genotype could be directed to these high-risk patients.
The American Rare Donor Registry and rare donor registries in other countries have identified and classified donors as hrB− or hrS−. These serologic designations are general classifications that are often inferior because there are no standardized antibody reagents and most antibodies that are called anti-hrB or -hrS do not have identical specificity. Apparent hrB− or hrS− phenotypes encompass multiple different Rh protein polymorphisms encoded by numerous RHCE*ce alleles. Therefore, it is important to note that not all donors with these phenotypes will be compatible and that, because they occur on very diverse Rh backgrounds, the complete RH genotype of the donor and patient must be considered to prevent further Rh sensitization.12,13
Currently in the American Rare Donor program administered by the American Red Cross, 87 hrB− or hrS− donors have been fully RH genotyped and 45 different RH genotypes were found, confirming the heterogeneous nature of RH genes. Several alloimmunized patients are currently supported with genetically matched donors with excellent results.38 However, the limited number of RH-genotyped donors in the ARDP currently precludes providing donor units to all but the highly immunized.
Conclusions
Sickle cell disease affects approximately 80,000 people in the United States, but this number represents only a fraction of those affected globally. Blood transfusion therapy remains the primary treatment for SCD. Indications for acute and chronic transfusion therapy continue to expand and include acute anemia, primary and secondary prevention of stroke, acute chest syndrome, splenic sequestration, pulmonary hypertension, priapism, and pre-operative preparation. Transfusion therapy is predicted to increase following the stroke prevention trials, STOP 1 and STOP 2, which demonstrated that prophylactic transfusion prevents stroke in children with sickle cell anemia who have abnormalities on transcranial Doppler (TCD) ultrasound and that discontinuation of transfusion therapy results in a high risk of reversion to abnormal TCD velocities and subsequent stroke.39,40 With transfusion therapy predicted to increase, and with the availability of oral iron chelation agents, approaches to minimize RBC alloimmunization are crucial to reduce complications and improve therapy.
RH genes in patients with SCD are diverse and are associated with Rh alloimmunization, in spite of transfusion with donor units antigen-matched for conventional Rh antigens. Routine serologic typing of their RBCs cannot distinguish these variant RH gene products from the conventional antigens, which may result in transfusions that are not truly Rh-matched. Subsequent Rh alloimmunization has the potential to compromise red cell survival. These findings emphasize the current shortcomings of serologic reagents, which detect only five primary antigens common in Caucasians, and suggest that RH “genetic-matching” would be a superior approach. RH genotyping would identify patients who are homozygous for variant alleles and at risk for production of alloantibodies to high prevalence Rh antigens and, when partnered with RH genotyping of donors, would allow selection of units that could eliminate the risk of Rh alloimmunization.
Comparison of antibodies present in patients with sickle cell disease (SCD) receiving C-, E-, and K-matched donor units from minority donors (Group A) and patients with SCD receiving random donor units (Group B).
| . | Group A (n = 20) . | Group B (n = 26) . | |
|---|---|---|---|
| *Often identified as apparent autoantibodies. | |||
| Total number of antibodies | 31 | 108 | |
| Common Rh alloantibodies | 0 | 22 | |
| 18 anti-E | |||
| 4 anti-C | |||
| Complex Rh antibodies* | 22 | 30 | |
| 11 anti-D (D+ patients) | 4 anti-D (D+ patients) | ||
| 8 anti-e (e+ patients) | 6 anti-e (e+ patients) | ||
| 3 anti-C (C+ patients) | 20 anti-C or -Ce (C+ patients) | ||
| Other antibodies | 9 | 56 | |
| 2 anti-Jkb | 8 anti-K | 1 anti-N | |
| 1 anti-Fya | 6 anti-S | 1 anti-Jsa | |
| 4 anti-M | 6 anti-Fya | 1 anti-Kpa | |
| 1 anti-N | 4 anti-Jkb | 1 anti-Yta | |
| 1 anti-Jsa | 2 anti-Jka | 1 anti-Lea | |
| 1 anti-M | 1 anti-Leb | ||
| 1 anti-Goa | |||
| RHalleles | |||
| Hybrid RHD-CE-D and RHCE*ces | 3 | 20 | |
| Only altered RHCE*ce | 11 | 6 | |
| Partial RHD and altered RHCE*ce | 9 | 14 | |
| . | Group A (n = 20) . | Group B (n = 26) . | |
|---|---|---|---|
| *Often identified as apparent autoantibodies. | |||
| Total number of antibodies | 31 | 108 | |
| Common Rh alloantibodies | 0 | 22 | |
| 18 anti-E | |||
| 4 anti-C | |||
| Complex Rh antibodies* | 22 | 30 | |
| 11 anti-D (D+ patients) | 4 anti-D (D+ patients) | ||
| 8 anti-e (e+ patients) | 6 anti-e (e+ patients) | ||
| 3 anti-C (C+ patients) | 20 anti-C or -Ce (C+ patients) | ||
| Other antibodies | 9 | 56 | |
| 2 anti-Jkb | 8 anti-K | 1 anti-N | |
| 1 anti-Fya | 6 anti-S | 1 anti-Jsa | |
| 4 anti-M | 6 anti-Fya | 1 anti-Kpa | |
| 1 anti-N | 4 anti-Jkb | 1 anti-Yta | |
| 1 anti-Jsa | 2 anti-Jka | 1 anti-Lea | |
| 1 anti-M | 1 anti-Leb | ||
| 1 anti-Goa | |||
| RHalleles | |||
| Hybrid RHD-CE-D and RHCE*ces | 3 | 20 | |
| Only altered RHCE*ce | 11 | 6 | |
| Partial RHD and altered RHCE*ce | 9 | 14 | |
Top. Diagram of theRHlocus. The figure illustrates the inverted orientation of the RHD and RHCE genes and the deletion of RHD associated with the Rh-negative RBC phenotype. Bottom. Predicted 12-transmembrane model of the RhD and RhCE proteins in the erythrocyte membrane. The amino acid differences between RhD and RhCE are shown as circles. The zigzag lines represent the location of palmitoylation sites. Amino acid positions 103 and 226 on RhCE that are critical for C or c and E or e expression, respectively, are indicated.
Top. Diagram of theRHlocus. The figure illustrates the inverted orientation of the RHD and RHCE genes and the deletion of RHD associated with the Rh-negative RBC phenotype. Bottom. Predicted 12-transmembrane model of the RhD and RhCE proteins in the erythrocyte membrane. The amino acid differences between RhD and RhCE are shown as circles. The zigzag lines represent the location of palmitoylation sites. Amino acid positions 103 and 226 on RhCE that are critical for C or c and E or e expression, respectively, are indicated.
Top. Diagram of the conventionalRHDandRHCEgenes. The coding regions consist of 10 exons, depicted as white and grey boxes, respectively. Bottom. Diagram of some RH genes commonly found in African ethnic groups. Shown are the RHD-RHCE gene that encodes altered C antigen, some examples of RHD encoding partial D (DIII, DAU, and DAR), and some examples of RHCE with mutations encoding altered or partial e that often complicate transfusion in sickle cell patients.
Top. Diagram of the conventionalRHDandRHCEgenes. The coding regions consist of 10 exons, depicted as white and grey boxes, respectively. Bottom. Diagram of some RH genes commonly found in African ethnic groups. Shown are the RHD-RHCE gene that encodes altered C antigen, some examples of RHD encoding partial D (DIII, DAU, and DAR), and some examples of RHCE with mutations encoding altered or partial e that often complicate transfusion in sickle cell patients.
Disclosures Conflict-of-interest disclosures: CMW is a consultant for Immucor. STC declares no competing financial interests. Off-label drug use: None disclosed.
References
Author notes
Division of Hematology, The Children’s Hospital of Philadelphia, Philadelphia, PA
American Red Cross, Philadelphia, PA; University of Pennsylvania, Department of Pathology and Laboratory Medicine, Philadelphia, PA