Abstract
Treatment response in patients with acute lymphoblastic leukemia (ALL) is best assessed using assays for minimal residual disease (MRD). The degree of leukemia cytoreduction and MRD clearance is determined by the collective influence of multiple factors. Some of these variables are features of the leukemic cells, such as expression of genes that regulate their susceptibility to cytotoxic drugs and their propensity to undergo apoptosis. Gene profiles depend, in turn, on the cell of origin for leukemic transformation, the type of underlying genetic abnormalities and/or epigenetic regulatory mechanisms. Another set of variables is related to the host, such as age and polymorphisms in genes that metabolize drugs, which together with pharmacologic variables, such as drug pharmacodynamics and drug interactions, influence treatment response. Finally, the bone marrow microenvironment where leukemic cells reside can participate in the generation of drug resistance. Altogether, these variables determine treatment outcome in each patient. Full knowledge of the molecular features associated with treatment response is required for precise leukemia prognostication and monitoring, and can provide clues to useful targets for novel therapies.
Clinical Significance of Molecular Measurements of Treatment Response
Better understanding of the unique molecular characteristics of leukemic cells has led to novel ways of detecting morphologically occult leukemic cells (i.e., minimal residual disease, MRD). Leukemia-associated molecular features include clonally rearranged immunoglobulin and T-cell receptor genes, chromosomal abnormalities and fusion transcripts amplifiable by polymerase chain reaction (PCR) as well as abnormalities in protein expression detectable by flow cytometry.
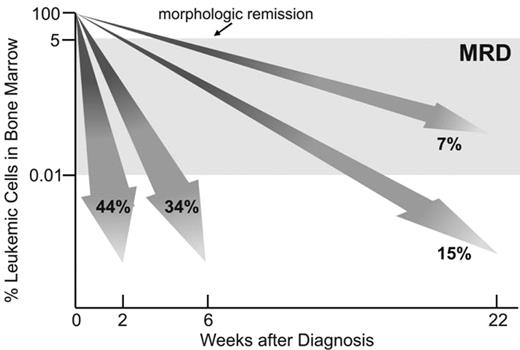
Studies of MRD in children with acute lymphoblastic leukemia (ALL) have demonstrated that response to initial therapy is heterogeneous. In trials at St. Jude Children’s Research Hospital, 44% of patients achieved MRD negativity on day 19, that is, less than 0.01% leukemic cells were detectable among bone marrow mononuclear cells (Figure 1 ). The remaining patients needed further treatment to achieve a similar level of leukemia cytoreduction but some stayed MRD-positive. The clinical relevance of MRD levels during the early phases of therapy was demonstrated by their strong correlation with relapse rates and overall treatment outcome.1–3 The finding that MRD is the strongest predictor of outcome in childhood ALL has been confirmed and extended by recent studies. In an update of their initial observations, investigators of the International Berlin-Frankfurt-Muenster (I-BFM) group reported a 10-year event-free survival of 93% for patients in the low-risk group (defined by negative MRD on both days 33 and 78; 43%) versus 16% for the high-risk group (defined by MRD levels of 0.1% or higher at both time points; 15%); 43% of the patients were deemed to have intermediate–risk ALL and had a 10-year event free survival rate of 74%.4 Investigators of the Dana-Farber Cancer Institute ALL Consortium studied MRD in 284 children with B-lineage ALL. The 5-year risk of relapse was 5% in 176 children with no detectable MRD at the end of remission induction and 44% in the 108 children with detectable MRD.5 In this study, an MRD cut-off level of 0.1% was found to be the one that best predicted 5-year relapse hazard: 72% for patients with higher levels of MRD and 12% for those with lower levels. Investigators of the Children’s Oncology Group (COG) monitored MRD in peripheral blood collected on day 8 and in bone marrow collected on day 29 (end of remission induction therapy) in 2143 children with B-lineage ALL.6 The presence of MRD (0.01% or higher) at either interval predicted a poorer outcome, and MRD levels in the day 29 bone marrow were the strongest prognostic factor in this series.
The prognostic significance of MRD has also been studied in adult patients with ALL, although less extensively than in children. Bruggeman et al7 monitored MRD in 196 adult patients with standard-risk ALL. They found that 10% of patients who had a rapid MRD decline to less than 0.01% on day 11 and day 24 had a 3-year relapse rate of 0%; 23% of patients had MRD of 0.01% or higher that persisted until week 16 and had a relapse rate of 94%. The remaining patients had a relapse rate of 47%. Raff et al8 analyzed bone marrow samples of 105 patients in hematologic remission who had completed first-year chemotherapy, and were MRD negative in previous tests. They found that 28 patients converted to MRD positivity and that 17 of 28 had relapsed at the time of the report with a median time from molecular to clinical relapse of 9.5 months. Of the remaining 77 patients who maintained MRD-negativity only 5 had relapsed.
Because molecular measurements of treatment response are considerably more powerful than traditional morphologic monitoring, they are being incorporated to guide therapy in many protocols. The specific timepoints at which MRD testing is most informative depend on the treatment regimen. In the recently completed Total XV study for newly diagnosed children with ALL at our institution, remission induction therapy was intensified for patients who had MRD 1% or higher on day 19 of remission induction therapy; postremission therapy was intensified for standard-risk patients who had MRD 0.01% or higher on day 46.9 Moreover, any patient with MRD 1% or higher on day 46, or 0.1% or higher during continuation therapy was considered as a candidate for allogeneic hematopoietic stem cell transplantation (HSCT). The BFM group uses MRD levels on days 33 and 78 as a guide for treatment intensification.4
Early clearance of MRD indicates a high chemosensitivity of the leukemic clone and was associated with an excellent overall outcome in correlative studies.10 On the basis of this observation and considering that in past trials nearly half of children with ALL could be cured with therapy less intensive than that of today, we hypothesize that patients who achieve MRD negativity after 2 to 3 weeks of remission induction chemotherapy can be cured with less intensive therapy. The need for a reduction in treatment intensification is particularly pressing in developing countries, where contemporary therapies for childhood ALL may have unacceptably high rates of toxicity. To this end, a protocol that incorporates reduction in treatment intensity for patients with negative MRD in bone marrow on day 19 as determined by a simplified flow cytometric assay10 has been implemented in Recife (Brazil).
In addition to measuring early response to chemotherapy, MRD assays have several other applications in the clinical management of patients with ALL (Table 1 ). For example, they can herald impending relapse, thus accelerating the planning of salvage therapy and/or HSCT. Since the risk of relapse after HSCT is strongly related to levels of MRD before transplant,11 MRD measurements can also be used to determine the timing of HSCT. MRD measurements post-HSCT can be used to guide the administration of donor lymphocyte infusions or other agents. Finally, in patients who relapse and achieve a second remission, MRD assays can be used to guide the selection of optimal post-remission treatment (i.e., chemotherapy versus HSCT).
Genetic Subtypes of ALL and Treatment Response
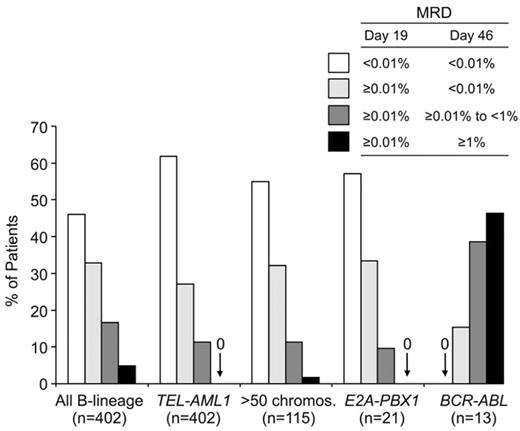
Among both children and adults with B-lineage ALL, those with the t(9;22)/BCR-ABL, t(4;11)/MLL-AF4 fusion, and hypodiploidy (< 44 chromosomes per leukemic cell) generally have a poorer treatment outcome, whereas hyperdiploidy (> 50 chromosomes), TEL-AML1 fusion, and trisomy 4, 10, and 17 are associated with a favorable prognosis.9,12 In a recent analysis of 706 children with B-lineage ALL enrolled in four consecutive treatment protocols at St Jude Children’s Research Hospital from 1991 to 2006, those with hyperdiploidy (> 50 chromosomes), TEL-AML1, and t(1;19)/E2A-PBX1 had the most favorable outcome, whereas those with the t(9;22)/BCR-ABL or t(4;11)/MLL-AF4 had a very poor outcome.9 Among 1522 adult patients enrolled on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial, those with at(9;22)(q34;q11), t(4;11)(q21;q23), t(8;14)(q24.1;q32), complex karyotype, or low hypodiploidy/near triploidy all had lower event-free and overall survival rates, whereas those with high hyperdiploidy or a del(9p) had a significantly better outcome.13 Studies that related MRD levels during remission induction therapy to molecular subtypes of ALL demonstrated that the impact of the molecular abnormalities is already exerted during the initial phases of therapy. Thus, in children with B-lineage ALL, MRD rates are particularly high in BCR-ABL–positive ALL and particularly low in TEL-AML1–positive ALL (Figure 2 ).
Among patients with T-lineage ALL, there is some suggestion that activating mutations of the NOTCH1 gene, found in around 50% of cases, are associated with a favorable prognosis in childhood cases,14 but an unfavorable outcome in adults.15 Paietta et al16 reported a unique subset of adult T-lineage ALL cases that coexpress CD117/KIT and cytoplasmic CD3. Activating mutations in the FLT3 gene were found in each of 3 cases that were analyzed, but not in 52 other adult T-ALL samples from the same series that lacked CD117 expression. We recently identified a subset of childhood T-ALL cases that represent the leukemic counterpart of early thymic precursors and have a dismal outcome (E. Coustan-Smith, C. Mullighan, D. Campana et al, manuscript submitted).
Discovery of Genes Associated with Treatment Response
Several studies used in vitro drug sensitivity testing to identify genes associated with drug resistance. Holleman et al17 studied leukemic lymphoblasts from 173 children and identified a set of genes associated with resistance to prednisolone, vincristine, asparaginase and daunorubicin that was significantly associated with treatment outcome in two separate cohorts. The most predictive genes were involved in a variety of cellular functions not immediately linkable to cellular drug resistance; other genes associated with cell proliferation, drug metabolism or apoptosis were associated with in vitro responses to individual drugs, but for most genes the statistical association was not sufficiently robust to warrant their designation as strong predictors of clinical outcome. Lugthart et al18 determined in vitro drug sensitivity of ALL cells in 441 patients and used a genome-wide approach to identify 45 genes differentially expressed in ALL exhibiting crossresistance to prednisolone, vincristine, asparaginase, and daunorubicin. The expression of these genes discriminated treatment outcome in two independent patient populations, identifying a subset of patients with a markedly inferior outcome. Also in this study, the function of the most discriminating genes was not obviously linked to drug resistance.
Holleman et al19 also investigated whether genes encoding key regulators of apoptosis were related to in vitro drug resistance and treatment outcome in ALL. They found that increased expression of the anti-apoptotic gene MCL1 was significantly associated with prednisolone resistance, whereas that of the proapoptotic gene HRK was associated with increased asparaginase sensitivity. However, higher expression of another proapoptotic gene, BCL2L13, was associated with a higher resistance to asparaginase and inferior treatment outcome, demonstrating the complexity of reconciling statistical association with molecular functions.
Other investigators used treatment response in vivo as a parameter to identify genes associated with drug resistance. Sorich et al20 performed gene expression studies in 161 children with ALL and related the findings to response to initial methotrexate treatment. They identified 48 genes and 2 cDNA clones whose expression was significantly related to the reduction of circulating leukemia cells after treatment, a finding that was validated in an independent cohort and was predictive of long-term disease-free survival. Genes significantly associated with response included those involved in nucleotide metabolism, cell proliferation, apoptosis, and DNA replication and repair.
Cario et al21 compared gene expression profiles of lymphoblasts in 21 B-lineage ALL patients with high MRD and 30 with low MRD enrolled in the BFM ALL-2000 protocol; leukemic cells in all patients lacked known genetic abnormalities predictive of outcome, but the expression of several genes was strongly associated with MRD; those with low expression in high-MRD cases were predominantly associated with cell-cycle progression and apoptosis. We analyzed gene expression of diagnostic lymphoblasts from 189 children with ALL and compared the findings with MRD on day 46 of remission induction treatment.22 After excluding genes associated with genetic subgroups, we identified 17 genes that were significantly associated with MRD. Low levels of one of the genes, caspase 8–associated protein 2 (CASP8AP2), were associated with higher MRD and predicted a lower event-free survival and a higher rate of leukemia relapse in a separate cohort of 99 patients. The reported proapoptotic role of CASP8AP2 fits well with these results and with the finding that higher CASP8AP2 expression was associated with a greater propensity of leukemic lymphoblasts to undergo apoptosis.22
Taking advantage of the same gene expression database, we also studied the relation between gene expression at diagnosis and MRD results obtained on day 19 of remission induction treatment.23 We identified 348 probe sets that were overexpressed in diagnostic samples from patients with MRD on day 19, and 326 that were under-expressed. Forty of these genes predicted leukemic relapse in the independent patient cohort and 14 showed independent prognostic significance. The function of some of the genes associated with MRD on day 19 and relapse hazard treatment in this study can be plausibly linked to the treatment that the patients received in the first 2 weeks of remission induction therapy. For example, lower expression of TOP2A, the target of daunorubicin activity, was associated with a higher MRD and risk relapse. Lower levels of BUB3, MAD2L1 and NUSAP1, which participate in mitotic spindle assembly, were expressed at lower levels in patients with MRD and an inferior outcome, consistent with the spindle poison activity of vincristine. Other genes that were underexpressed in cases with a higher MRD and relapse hazard, such as CCNB2, CDC2, and CKS1B, are essential for cell-cycle progression, a function with direct relevance to the drug sensitivity of leukemic cells.
Some gene expression studies focused on specific subsets of ALL. Juric et al24 studied samples from 37 adult patients from the Medical Research Council UKALL XII/Eastern Cooperative Oncology Group E2993 trial with BCR-ABL–positive ALL and 17 with BCR-ABL-negative ALL. They identified 27 genes, many forming a network involved in cellular differentiation, that correlated with overall survival in BCR-ABL-positive cases. Chiaretti et al25 examined gene expression profiles of 33 adult patients with T-ALL identified a set of 30 genes that was highly expressed in leukemic cells from patients who achieved complete remission; 19 genes were differentially expressed in patients who either had a relapse or remained in remission. A model based on the expression of 3 of these genes (AHNAK, TTK and CD2) was predictive of remission duration. Ferrando et al26 found that HOX11 expression was significantly associated with a favorable prognosis in children with this leukemia subtype, while expression of TAL1, LYL1, or HOX11L2 conferred a worse response to treatment. These authors also found that adult T-ALL patients whose lymphoblasts expressed TLX1 had significantly better survival rates.27 Baldus et al28 determined ERG and BAALC mRNA expression by real-time reverse transcriptase PCR in 153 adults with T-ALL and found that patients with low expression of both ERG and BAALC (41%) had the most favorable outcome. Within the subgroup of thymic T-ALL, high ERG and the presence of HOX11L2 were independent predictors of an adverse outcome.
Conceivably, the identification of genes that are commonly associated with treatment response in different studies will help focussing on the most relevant cellular functions and molecular pathways in ALL. Table 2 lists genes found to be associated with drug resistance in more than one of four studies of childhood ALL examined.17,18,21,23 Interestingly, low expression of one of the genes, TTK, was also found to be associated with inferior response in adult T-ALL.25 Moreover, in adult patients with follicular lymphoma who received combination chemotherapy that included vincristine, prednisone and an anthracycline, lower expression of CDC2 and CKS1B was associated with an inferior treatment response,29 in agreement with studies in childhood ALL.17,23
Polymorphisms in Drug-Metabolizing Genes and Treatment Response
Variation in the expression and function of genes involved in drug metabolism has been associated with treatment response in children with ALL.30 For example, polymorphisms and the activity of thiopurine methyltransferase, an enzyme that inactivates thiopurines, influence response to this class of drugs.30,31 Thus, when treated with conventional doses of thiopurines, patients with heterozygous deficiency (~10%) or homozygous deficiency (~0.03%) are at risk of developing serious hematopoietic toxic effects.30 The enzyme deficiency also confers a higher risk of developing therapy-related acute myeloid leukemia and radiation-induced brain tumors in patients receiving thiopurines.30 On the other hand, patients with high levels of enzyme activity might be at greater risk of relapse due to a decrease in exposure of leukemic cells to active drug metabolites.31
Rocha et al32 found that treatment outcome in 246 children with ALL was related to 16 polymorphisms in genes involved in the pharmacodynamics of antileukemic drugs. Among the 130 patients treated in the higher-risk arms of the Total XIII St Jude study, those with the glutathione S-transferase (GSTM1) non-null genotype had a greater risk of hematologic relapse, which was further increased by the thymidylate synthetase (TYMS) 3/3 genotype. Importantly, expression of GSMT1 and TYMS in ALL blasts was lower in those with low-activity genotypes. de Jonge et al33 studied whether polymorphisms in genes involved in folate metabolism affected methotrexate sensitivity in vitro in 157 childhood ALL cases. Patients with the methylenetetra-hydrofolate reductase (MTHFR) 1298AC variant and those with the methionine synthase reductase (MTRR) 66 G-allele had a decreased sensitivity. Davies et al34 studied polymorphisms of 16 genes with functions related to treatment response in 1197 children enrolled in Pediatric Oncology Group therapy protocols 9904, 9905 and 9906, and compared the findings to MRD levels on day 8 in blood and on day 28 in bone marrow. After adjusting for known prognostic features, these investigators found that G allele of a common polymorphism in the chemokine receptor 5 (CCR5) gene was associated with a better MRD clearance than the A allele, although the molecular mechanisms underlying this association are unclear.
Role of the Microenvironment
Leukemic cells that infiltrate tissues such as the central nervous system (CNS) are exposed to lower drug concentrations and may persist even after intensive chemotherapy, causing relapse. It is unclear whether CNS infiltration is a random event or is directed by the expression of specific molecules in leukemic cells. Cario et al35 studied leukemic gene expression profiles from the bone marrow of 17 CNS-positive patients and 26 CNS-negative patients and found that interleukin-15 (IL-15) expression was consistently upregulated in leukemic cells of CNS-positive patients. In patients who were CNS-negative at diagnosis, IL-15 levels greater than the median were associated with subsequent CNS relapse, further suggesting that the propensity to infiltrate the CNS is related to this biologic feature of ALL cells.
The survival and growth of leukemic cells depends on the support provided by mesenchymal cells in the bone marrow.36 An emerging concept is that the bone marrow microenvironment can also regulate the response of leukemic cells to chemotherapy through a variety of mechanisms. For example, mice injected with Arf-null pre-B cells expressing p185 BCR-ABL die of ALL and respond poorly to imatinib therapy, but treatment response is markedly increased if cytokine signaling is abrogated, suggesting that host cytokines contribute to drug resistance.37 We studied the effect of the microenvironment on the sensitivity of ALL cells to asparaginase. ALL cells are particularly sensitive to asparagine and glutamine depletion caused by asparaginase because their capacity to produce their own asparagine supply is extremely low, due to their low expression of the asparagine synthetase gene (ASNS). Stams et al38 found that higher levels of ASNS expression were associated with greater resistance to asparaginase treatment in vitro and an inferior treatment outcome in patients with some but not all genetic subtypes of ALL. However, Fine et al39 reported that ASNS expression in ALL cells could not predict cellular responses to asparaginase in vitro and, according to Holleman et al,17ASNS was not one of the genes that best predicted resistance to asparaginase in vitro. Indeed, we found that levels of ASNS expression in 288 children with ALL were not related to in vivo responses to therapy that included asparaginase. We observed, however, that ASNS expression levels in bone marrow-derived mesenchymal cells (which form the microenvironment where leukemic cells grow) were, on average, 20 times higher than those in ALL cells.36 Mesenchymal cells protected ALL cells from asparaginase cytotoxicity in coculture experiments. This protective effect correlated with levels of ASNS expression; downregulation by RNA interference decreased mesenchymal cell capacity to protect ALL cells from asparaginase, whereas enforced ASNS expression conferred enhanced protection. Asparagine secretion by mesenchymal cells was directly related to their ASNS expression levels, suggesting that increased concentrations of asparagine in the leukemic cell microenvironment constitute the underlying mechanism for the protective effects we observed.
Conclusions and Future Challenges
Molecular assessment of treatment response is increasingly used to guide therapeutic decisions. The available flow cytometry– and PCR-based MRD assays are reliable but quite complex and can be performed well only in highly specialized laboratories. Hence, it is important to further simplify MRD technologies so that they can be implemented widely. MRD studies can also provide a powerful tool to quickly assess the effectiveness of novel antileukemic agents and support innovative designs for Phase II studies.
The explosion of information about the genes that are associated with treatment outcome in ALL has fueled the formulation of new hypotheses about the molecular mechanisms underlying drug resistance. The information that has been gained on outcome prediction should be validated in prospective trials by including the most promising genes in dedicated leukemia gene expression or real-time PCR arrays, and/or adding antibodies that recognize the corresponding proteins to leukemia diagnosis panels. Mechanistic links need to be established between gene expression and drug resistance to identify the key pathways to be targeted by novel therapies. The clinical potential of this newly acquired information is well illustrated by the study of Wei et al,40 which used novel analytical methods to screen a database of drug-associated gene expression profiles for molecules whose profiles overlapped with a gene expression signature of glucocorticoid sensitivity/resistance in ALL cells. The screen indicated that the mTOR inhibitor rapamycin profile matched the signature of glucocorticoid sensitivity. The authors found that rapamycin would indeed induce glucorticoid sensitivity in leukemic cells via modulation of the antiapoptotic molecule MCL1.
A recent study using high-resolution, single-nucleotide polymorphism arrays and genomic DNA sequencing revealed deletion, amplification, point mutation and structural rearrangement in genes encoding key regulators of B lymphocyte development and differentiation in 40% of B-lineage ALL cases.41 The PAX5 gene was the most frequent target of somatic mutation, being altered in 32% of cases, resulting in reduced protein levels or the generation of hypomorphic alleles. Deletions were also detected in the TCF3 (E2A), EBF1, LEF1, IKZF1 (IKAROS) and IKZF3 (AIOLOS) genes. Although the prognostic significance of these findings remains to be determined, the study illustrates well the potential of contemporary technologies to uncover previously unnoticed molecular abnormalities in leukemic cells, at least some of which should regulate pathways that intersect with those determining cellular responses to chemotherapy.
Molecular measurements of treatment response—clinical applications.
| Application . | Action . |
|---|---|
| Abbreviations: MRD, minimal residual disease; HSCT, hematopoietic stem cell transplantation; DLI, donor lymphocyte infusion | |
| Detect poor early treatment response | Treatment intensification |
| Detect good early treatment response | Treatment deintensification |
| Detect relapse | Treatment intensification; prepare for HSCT |
| Measure MRD before HSCT | Optimize timing of HSCT |
| Measure MRD after HSCT | Modulate immunosuppression, DLI, etc. |
| Measure MRD at the end of experimental remission induction protocol | Apply MRD-based stopping rule |
| Evaluate novel anti-leukemic agents | Novel design of Phase II studies |
| Application . | Action . |
|---|---|
| Abbreviations: MRD, minimal residual disease; HSCT, hematopoietic stem cell transplantation; DLI, donor lymphocyte infusion | |
| Detect poor early treatment response | Treatment intensification |
| Detect good early treatment response | Treatment deintensification |
| Detect relapse | Treatment intensification; prepare for HSCT |
| Measure MRD before HSCT | Optimize timing of HSCT |
| Measure MRD after HSCT | Modulate immunosuppression, DLI, etc. |
| Measure MRD at the end of experimental remission induction protocol | Apply MRD-based stopping rule |
| Evaluate novel anti-leukemic agents | Novel design of Phase II studies |
Genes whose expression was associated with drug resistance in ALL in more than one study.
| Probe set ID* . | Gene name . | Gene symbol . | Main biological processes . | References† . |
|---|---|---|---|---|
| *In Affymetrix U133 GeneChip | ||||
| †References in which gene expression was associated with MRD or drug sensitivity in vitro | ||||
| Genes overexpressed in cases with drug resistance in vitro or in vivo | ||||
| 201376_s_at | Heterogeneous nuclear ribonucleoprotein F | HNRPF | RNA processing | 18, 23 |
| 205644_s_at | Small nuclear ribonucleoprotein polypeptide G | SNRPG | Spliceosome assembly, mRNA processing | 17, 23 |
| 208808_s_at | High-mobility group box 2 | HMGB2 | DNA replication, DNA repair, nucleosome assembly, regulation of transcription, phosphoinositide-mediated signaling | 18, 23 |
| 209760_at | KIAA0922 | KIAA0922 | 17, 23 | |
| 213911_s_at | H2A histone family, member Z | H2AFZ | Nucleosome assembly | 18, 23 |
| 218381_s_at | Sphingosine-1-phosphate lyase 1 | SGPL1 | Lipid metabolism, ceramide metabolism, apoptosis | 17, 23 |
| 218802_at | Coiled-coil domain containing 109B | CCDC109B | 17, 18, 23 | |
| Genes underexpressed in cases with drug resistance in vitro or in vivo | ||||
| 38710_at | OTU domain, ubiquitin aldehyde binding 1 | OTUB1 | Ubiquitin cycle | 17, 23 |
| 201088_at | Karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | KPNA2 | DNA recombination, microtubule process, cell cycle, protein import into nucleus | 17, 23, 17, 23 |
| 201694_s_at | Early growth response 1 | EGR1 | Regulation of transcription | 17, 23 |
| 201755_at | Minichromosome maintenance complex component 5 | MCM5 | DNA replication, regulation of transcription | 21, 23 |
| 201897_s_at | CDC28 protein kinase regulatory subunit 1B | CKS1B | Cell cycle | 17, 23 |
| 202499_s_at | solute carrier family 2, member 3 | SLC2A3 | Carbohydrate metabolism, transport | 18, 23 |
| 203362_s_at | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2L1 | Cell cycle | 21, 23 |
| 204822_at | TTK protein kinase | TTK | Protein phosphorylation, mitotic spindle organization, cell proliferation | 21, 22 |
| 205193_at | v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | MAFF | Regulation of transcription | 18, 23 |
| 209795_at | CD69 | CD69 | Signal transduction | 18, 23 |
| Probe set ID* . | Gene name . | Gene symbol . | Main biological processes . | References† . |
|---|---|---|---|---|
| *In Affymetrix U133 GeneChip | ||||
| †References in which gene expression was associated with MRD or drug sensitivity in vitro | ||||
| Genes overexpressed in cases with drug resistance in vitro or in vivo | ||||
| 201376_s_at | Heterogeneous nuclear ribonucleoprotein F | HNRPF | RNA processing | 18, 23 |
| 205644_s_at | Small nuclear ribonucleoprotein polypeptide G | SNRPG | Spliceosome assembly, mRNA processing | 17, 23 |
| 208808_s_at | High-mobility group box 2 | HMGB2 | DNA replication, DNA repair, nucleosome assembly, regulation of transcription, phosphoinositide-mediated signaling | 18, 23 |
| 209760_at | KIAA0922 | KIAA0922 | 17, 23 | |
| 213911_s_at | H2A histone family, member Z | H2AFZ | Nucleosome assembly | 18, 23 |
| 218381_s_at | Sphingosine-1-phosphate lyase 1 | SGPL1 | Lipid metabolism, ceramide metabolism, apoptosis | 17, 23 |
| 218802_at | Coiled-coil domain containing 109B | CCDC109B | 17, 18, 23 | |
| Genes underexpressed in cases with drug resistance in vitro or in vivo | ||||
| 38710_at | OTU domain, ubiquitin aldehyde binding 1 | OTUB1 | Ubiquitin cycle | 17, 23 |
| 201088_at | Karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | KPNA2 | DNA recombination, microtubule process, cell cycle, protein import into nucleus | 17, 23, 17, 23 |
| 201694_s_at | Early growth response 1 | EGR1 | Regulation of transcription | 17, 23 |
| 201755_at | Minichromosome maintenance complex component 5 | MCM5 | DNA replication, regulation of transcription | 21, 23 |
| 201897_s_at | CDC28 protein kinase regulatory subunit 1B | CKS1B | Cell cycle | 17, 23 |
| 202499_s_at | solute carrier family 2, member 3 | SLC2A3 | Carbohydrate metabolism, transport | 18, 23 |
| 203362_s_at | MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2L1 | Cell cycle | 21, 23 |
| 204822_at | TTK protein kinase | TTK | Protein phosphorylation, mitotic spindle organization, cell proliferation | 21, 22 |
| 205193_at | v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | MAFF | Regulation of transcription | 18, 23 |
| 209795_at | CD69 | CD69 | Signal transduction | 18, 23 |
Molecular response to treatment in childhood acute lymphoblastic leukemia (ALL). Shown are percentages of patients enrolled in Total XIII, XIV and XV Studies at St Jude Children’s Research Hospital who were MRD-negative (< 0.01% leukemic cells in bone marrow) at sequential treatment intervals.
Molecular response to treatment in childhood acute lymphoblastic leukemia (ALL). Shown are percentages of patients enrolled in Total XIII, XIV and XV Studies at St Jude Children’s Research Hospital who were MRD-negative (< 0.01% leukemic cells in bone marrow) at sequential treatment intervals.
Molecular response in childhood B-lineage ALL according to genetic subtypes. Patients enrolled in Total XIII, XIV and XV Studies at St Jude Children’s Research Hospital were grouped according to MRD status on days 19 and 46 as shown. MRD levels in patients with BCR-ABL- and TEL-AML1-positive ALL were significantly different from that of patients lacking these genetic abnormalities.
Molecular response in childhood B-lineage ALL according to genetic subtypes. Patients enrolled in Total XIII, XIV and XV Studies at St Jude Children’s Research Hospital were grouped according to MRD status on days 19 and 46 as shown. MRD levels in patients with BCR-ABL- and TEL-AML1-positive ALL were significantly different from that of patients lacking these genetic abnormalities.
Disclosures Conflict-of-interest disclosure: The author declares no competing financial interests Off-label drug use: None disclosed.
Acknowledgments
This work was supported by grants CA60419, CA115422 and CA21765 from the National Cancer Institute, and by the American Lebanese Syrian Associated Charities (ALSAC).
References
Author notes
Departments of Oncology and Pathology, St. Jude Children’s Research Hospital, and Department of Pediatrics, University of Tennessee Health Science Center, Memphis, TN