Abstract
Burkitt lymphoma (BL), a tumor occurring in endemic, sporadic and AIDS-associated forms, is the classic example of a human malignancy whose pathogenesis involves a specific cellular genetic change, namely, a chromosomal translocation deregulating expression of the c-myc oncogene, complemented in many cases by the action of an oncogenic virus, the Epstein-Barr virus (EBV). Here we review recent work in two complementary areas of research: (1) on cellular genetic changes that occur in addition to the c-myc translocation in BL, in particular the capacity of p53/ ARF pathway breakage or of c-myc mutation to decouple the pro-proliferative effects of c-myc deregulation from its pro-apoptotic effects; and (2) on a postulated role for EBV in BL pathogenesis, through adopting restricted forms of virus latent gene expression that remain compatible with the c-myc–driven growth program but offer the tumor additional protection from apoptosis. We stress the many fundamental questions that remain to be resolved and, in that regard, highlight the general lessons that might be learned through understanding how two other infectious agents, malaria and HIV, dramatically enhance BL incidence.
Burkitt lymphoma (BL), first described by Denis Burkitt as an obscure tumor of African children,1 has since assumed paradigmatic status as the first human tumor whose pathogenesis could be linked both to an oncogenic virus, Epstein-Barr virus (EBV), and to the activation of a specific cellular oncogene, c-myc. How these factors complement one another in the oncogenic process, and what other factors play a role in disease pathogenesis, continues to fascinate virologists and oncologists alike.
Endemic, Sporadic and AIDS-Associated BL
Three epidemiologically distinct forms of BL are now recognized,2 and their salient features are summarized in Table 1 . The high-incidence “endemic” form typically presents as a jaw or abdominal tumor in children in areas of equatorial Africa and Papua New Guinea, where malaria is holoendemic, and is 100% EBV genome–positive. Elsewhere BL occurs in “sporadic” form, again mainly in children, at intermediate to low incidence and with different degrees of EBV association depending upon the area. Western countries show the lowest incidence rates and the weakest virus association, with only 15% to 20% tumors being EBV positive (+); by contrast, BL appears to be more common in other locations, for example, equatorial areas of Brazil, and EBV-association rates are correspondingly higher. It is therefore possible that chronic immune stimulation from other parasitic infections may also increase BL incidence to some degree and that this increase also preferentially involves EBV-associated disease. Remarkably, a third, adult form of the tumor, AIDS-BL, proved to be very common among HIV-infected individuals, often appearing as one of the first symptoms of AIDS; 30% to 40% of these tumors carry EBV. Classically, all BLs, irrespective of form or EBV status, carry a reciprocal translocation that places the c-myc gene under the control of either the heavy- or light-chain immunoglobulin (IgH or IgL) loci, leading to high-level c-myc expression. Such c-myc deregulation therefore appears to be the obligatory defining feature of BL pathogenesis, often with EBV acting as a complementary oncogenic agent, and other infections such as malaria or HIV, though not oncogenic per se, somehow accentuating lymphoma risk.
Histologic Diagnosis of BL
All forms of BL show a similar histologic appearance, with a malignant population of round monomorphic B cells, interspersed with macrophages that give the tumor histology a “starry sky” pattern (Figure 1; see Color Figures, page 514). The malignant cells also display an unusually high level of proliferation, indicated by a Ki67 score of greater than 95%, and express combinations of markers (BCL6+, CD10+, CD38+) that are characteristic of germinal center B cells,2 i.e., the stage at which antibody responses mature through somatic hypermutation of immunoglobulin [Ig] genes and Ig class switching. In practice, however, the histologic distinction between classical BL and certain presentations of diffuse large B cell lymphoma (DLBCL) can be difficult to draw. However, distinguishing these pathogenetically distinct entities is important clinically since, compared with DLBCL, the successful treatment of BL requires a much more intensive chemotherapeutic regimen.
Recent Advances in the Molecular Diagnosis of BL
In light of the above, two recent studies have used microarray technology in an attempt to identify gene expression signatures that distinguish BL, and “Burkitt-like” lymphomas, which have genetic abnormalities and immunophenotype of BL but have atypical morphologic features, from DLBCL.3,4 In each study, more than 200 aggressive B-cell lymphomas, previously diagnosed histologically by a panel of expert pathologists, were analysed by gene expression array; in addition, the genetic complexity of these same tumors was assessed either by fluorescent in situ hybridization (FISH) analysis to detect c-myc translocations or by array-based comparative genomic hybridization to detect other genetic changes. This expression profiling clearly identified a set of tumors with the molecular signature of BL (mBL); while this signature does indeed overlap with that of normal germinal-center B cells, it is distinct from DLBCL by including high expression of c-myc itself and of a number of c-myc–activated genes. Importantly, the molecularly defined mBL tumor set encompassed all those tumors with a classical BL histology, some but not all of the tumors diagnosed as “Burkitt-like,” and even a few cases diagnosed as DLBCL by histopathology.
The powerful combination of gene expression array with genetic analysis made it clear that most cases of molecularly defined BL carried a c-myc/Ig gene translocation, and relatively few if any additional genetic changes detectable by comparative genome hybridization. This emphasizes the dominant influence of this translocation on the BL phenotype. Yet interestingly, although high c-myc expression is an integral part of the mBL signature, a few molecularly defined BLs lacked such a translocation, implying that in rare cases the tumor can arise as a result of c-myc deregulation by another route. As to the implications of these studies for BL diagnosis, currently it is not practical or cost-efficient to use microarray analysis as a routine diagnostic tool. However, the array work has identified additional features of the mBL phenotype, such as TCL1 and BCL6 expression in the absence of CD44 and MUM1, that can be assayed by immunohistochemistry and can augment markers currently in use.
It is important to note that both of these recent array-based studies were conducted on collections of lymphomas from Western countries and so, by definition, were focusing on sporadic forms of BL. While EBV status was not recorded in the published reports, it is likely that only a minority of the molecularly defined BLs were EBV+. An important priority for future work will be to determine how closely endemic BL accords to the molecular signature of the sporadic disease and, indeed, whether there are specific changes in cellular gene expression associated with the presence of EBV.
Cellular Genetic Change and BL Pathogenesis
C-myc translocation and the effects of c-myc overexpression
C-myc is a transcription factor involved in many cellular processes including growth, proliferation and apoptosis. In accordance with this global role, c-myc expression is tightly regulated and immediately sensitive to external stimuli. In normal cells, c-myc’s pro-proliferative effects, exemplified by upregulation of cyclins D and E and down-regulation of the negative regulator p27, are counter balanced by apoptotic checkpoints, such that c-myc over-expression activates the p53 program through the nucleolar tumor suppressor ARF. Deregulation of c-Myc has been implicated in the pathogenesis of a number of human cancers and, in different tumor contexts, can occur through gene translocation or amplification, mRNA stabilization, enhanced translation or protein stabilization (reviewed in Dang et al5).
In the case of BL, c-myc deregulation typically results from one of three reciprocal translocations, involving the IgH gene (t(8;14)) in around 80% of cases and the kappa (t(8;22)) or lambda (t(2;8)) IgL genes in the remainder. Where studied, translocations into the light chain loci invariably place the whole c-myc gene some distance from the IgL sequence. By contrast, the anatomy of the t(8:14) translocation falls into one of two broad patterns that tend to correlate with the epidemiologic form of the tumor. Thus, in endemic BL the breakpoint in c-Myc typically occurs more than 100 kb upstream of the first coding exon, and the breakpoint in IgH occurs in the joining region. By contrast, in sporadic BL and AIDS-BL, the breakpoint in c-Myc often occurs between exons 1 and 2, and the breakpoint in IgH occurs in the switch region. The potential significance of these differences, in terms of the timing of the translocation during B-cell development, has been widely debated, but the issue remains unresolved.6 –8
There have been several attempts to recapitulate BL in mouse models using Ig-myc fusion transgenes.9,–11 In early work, transgenic mice carrying a wild-type c-myc gene fused to an IgH enhancer developed tumors, but these were of pre-B cell origin and did not resemble classical BL.9 More interestingly, later work used a transgene that reconstructed the c-myc/IgL enhancer translocation from a naturally occurring case of BL, where the c-myc gene itself carried mutations. This produced mouse B-cell tumors with a histology and cell phenotype closely resembling BL, but which were different in having a naïve (nonmutated) Ig genotype rather than the hypermutated Ig genotype typical of the human tumor.9 –11 These findings with a naturally occurring gene fusion, and earlier reports of c-myc sequence changes in BL, suggest that c-myc gene mutations, arising as a direct result of the translocation itself and/or through inappropriate targeting by the somatic hypermutation machinery that is naturally activated in germinal center cells, render the overexpressed c-myc protein more oncogenic.
The precise mechanisms whereby c-myc drives BL tumorigenesis are still not fully understood. Some insights have come from an in vitro reconstruction system in which a lymphoblastoid cell line (LCL), established from normal human B cells using an EBV strain whose transforming function is estrogen-dependent, was transfected with either a constitutive or a tetracycline-regulatable c-myc gene.12,–14 Overexpression of c-myc in these cells renders cell growth independent of EBV and, very interestingly, induces downregulation of LCL-associated cell adhesion and activation molecules and concomitant upregulation of BL-associated germinal center markers such as CD10 and CD38.13,15 Such findings reinforce the arguments from transgenic mouse work that c-myc overexpression in B cells can impose a germinal center–like phenotype even though, ironically, normal germinal center cells do not express c-myc;7 the findings also caution against making the too easy assumption that BL, because it displays a germinal center phenotype, is necessarily of germinal center cell origin. Interestingly, the in vitro reconstruction model also showed that c-myc expression imposed on LCL cells a non-immunogenic phenotype typical of BL itself, reducing the expression of HLA molecules and components of the antigen-processing pathway, downregulating many genes involved in the NF-κB response, and impairing the cellular response to type I interferons.13,16,17 This raises the possibility of another role for c-myc in BL pathogenesis, in aiding the tumor’s capacity to evade host immune controls.
More recent studies to determine the role of c-myc in tumorigenesis have used microarray technology to look at the genomic binding sites of c-myc in BL cells. Interestingly, c-myc was found bound to nearly 15% of gene loci tested, consistent with a global role for the protein in determining the malignant BL phenotype.18 Notable among c-myc target genes identified in this study were several involved in driving apoptosis, reflecting the known link between c-myc expression and apoptosis susceptibility seen in normal cells. Indeed, there is evidence both from histologic analysis of tumor biopsies and from tumor explantation in vitro that BL, though a rapidly proliferating tumor, nevertheless also suffers from high rates of apoptosis.
Lesions in the p53 and RB pathways
While c-myc deregulation appears to be an essential feature of BL, additional genetic and epigenetic alterations affecting the p53 and RB pathways have been detected in BL tumors or derived cell lines and are thought to be important in disease pathogenesis.19
Specifically, it has been reported that 30% of endemic BL tumors and up to 70% of long-established BL lines carry mutations in p53, often clustering around the core DNA binding and activation domain and thus preventing p53-mediated apoptosis and cell cycle arrest (reviewed in Lindstrom and Wiman19). Furthermore, those endemic BLs carrying a wild-type p53 gene frequently carry genetic alterations at other sites in the p53 and RB pathways, such as overexpression of the p53-negative regulator MDM2, silencing of p16INK4A through promoter methylation and deletion and, in a minority of cases, inactivation of the tumor suppressor p14ARF through homozygous deletion.19 The situation in sporadic BL is similar, with 55% of cases carrying a disrupted ARF-MDM2-p53 pathway due either to a mutation in the p53 gene or, more often, to MDM2 overexpression.20 Furthermore, of the clonal pre-B and B-cell lymphomas arising in mice with a c-myc/IgH enhancer transgene, some 28% carried p53 mutations and 24% had p14ARF/p16INK4A deletion or overexpressed MDM2.21 This again suggests that disruption of these pathways can complement c-myc–driven malignant change. Note, however, that BL cells with known impairment of the p53 pathway are also highly prone to apoptosis, indicating their continued susceptibility to death via one or more p53-independent routes.
Although the pRB/p105 pathway of negative growth regulation appears to be functional, lesions in a second regulatory pathway have been observed in several BL lines; these stem from mutations in the RBL2/p130 gene that impair nuclear localization of its protein product.22,23 Interestingly, tumor status with respect to p130 and to p107, a growth-related protein whose expression inversely correlates with that of p130 in normal cells, tends to differ in the three forms of BL. Thus, endemic BLs generally display cytoplasmic p130 and low-level nuclear p107, and sporadic BLs have low level nuclear p130 but high level nuclear p107, while AIDS-BLs show high nuclear expression of both proteins.23,24 The significance of these findings is not yet understood, though the situation in AIDS-BL may reflect an ability of the soluble TAT protein (derived from HIV-infected T cells) to bind to and inactivate p130,24,25 resulting in the de-repression of p107 and other pro-proliferative genes. While the relevance of p130 inactivation in BL pathogenesis remains to be determined, the results of a recent study, in which re-introducing wild-type p130 into endemic BL cells led to a G0–G1 cell-cycle block, are at least consistent with such a role.26
The effects of c-myc mutation
As described above, much work has focused on identifying additional genetic changes in BL that disrupt the balance between the dual effects of c-myc expression, proliferation and apoptosis. However, it is becoming clear that c-myc itself can become mutated in such a way as to retain proliferative function but impair the apoptotic signal.27,–29 C-myc mutations in BL frequently affect the myc box I region, often at residues 57 or 58, and are thought to increase the stability of the myc protein by preventing the threonine 58 phosphorylation that targets the protein for proteasomal degradation. While in vitro assays comparing wild-type and mutant myc alleles for differences in tumorigenic potential and apoptosis have not been conclusive,30,31 recent in vivo work in a murine model has shed new light on the likely relevance of c-myc mutants in BL pathogenesis.29
In this study, mice were irradiated and their immune systems reconstituted with hematopoetic stem cells transduced with a retroviral vector carrying either wild-type or mutated (T58A or P57S) c-myc alleles. Expression of mutant c-myc greatly accelerated lymphoma development. However, this was not associated with any increased proliferative capacity of the transduced cells, nor did the resultant tumors show the mutations in p19ARF/p53 pathway genes that typically appear as complementary changes in the later-appearing lymphomas driven by wild-type c-myc expression. Instead, the mutant c-myc-induced tumors were distinguished from their wild-type counterparts by markedly reduced expression of the pro-apoptotic BH3-only protein Bim.29 Importantly, in another context, Bim, which binds to and inactivates the anti-apoptotic Bcl2 protein, has been shown to play a key role in c-myc–induced apoptosis via a p53-independent mechanism.32 Indeed, the use of genetically modified stem cells showed that the enhanced lymphomagenic capacity of mutant compared to wild-type c-myc alleles was dependent upon the presence of Bim; furthermore, unlike the wild-type allele, mutant c-myc was equally lymphomagenic in the presence or absence of p53.29
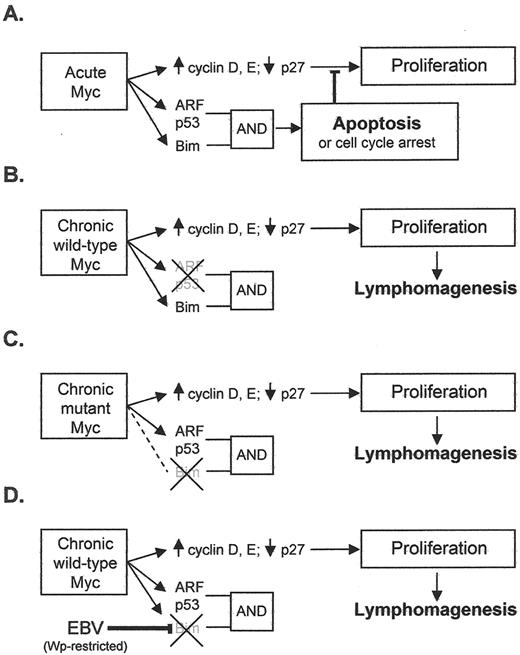
These findings support the view that, in the physiologic situation, acute c-myc expression activates genes involved in cell proliferation and also, as a counterbalance, genes of the p53/ARF pathway and the Bcl2-inhibitor Bim, which, though operating by different pathways, combine to deliver a cumulative apoptotic signal (Figure 2A 28). They further lead to the hypothesis that any changes that reduce this cumulative signal will predispose to c-myc–driven lymphomagenesis. A prediction of the hypothesis is that two features would distinguish BLs with a codon 57 or 58 mutation in the translocated c-myc allele from BLs with a wild-type allele: an over-representation of tumors lacking Bim expression and an under-representation of tumors with p53 mutation (see Figures 2B and C 28). This was indeed the case in the tumors analysed in that report.29
EBV and BL Pathogenesis
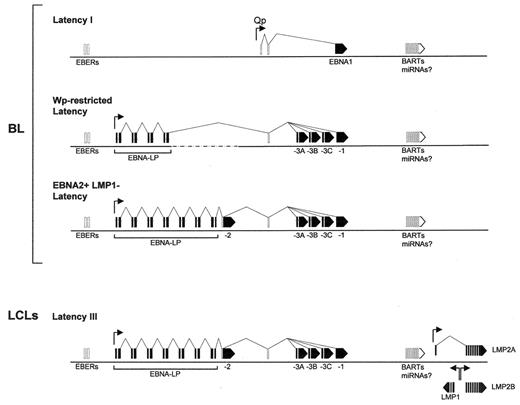
EBV, a gamma1-herpesvirus carried by the vast majority of individuals worldwide as a lifelong asymptomatic infection, nevertheless has growth-transforming potential and is aetiologically linked with a variety of B-cell, T/NK-cell and epithelial malignancies. However, use of the full growth-transforming program appears to be restricted to B cells33 and, indeed, infecting such cells in vitro leads to the outgrowth of permanent EBV+ LCLs. This growth-transforming program (called Latency III) involves expression of six EB nuclear antigens—EBNAs 1, 2, 3A, 3B, 3C and -LP—from one of two adjacent promoters Wp or Cp and two latent membrane proteins—LMPs 1 and 2—from their own promoters; in addition, BamHI A rightward transcripts (BARTS, a presumed source of the virally encoded BamHIA microRNAs) and the non-coding EBER transcripts are detectable here, as well as in all other forms of latency (see Figure 3 ). This same program is also seen in most cases of EBV-driven post-transplantation lymphoproliferative disease (PTLD) lesions to which immunosuppressed patients who have received transplants are particularly prone.2
By contrast, EBV+ BLs (whether endemic, sporadic or AIDS-associated) typically display more restricted forms of latency; indeed, it is clear from in vitro models that the full Latency III program is incompatible with the high-level c-myc expression that is the essential determinant of the BL phenotype.15 Thus, most BLs carry EBV as a Latency I infection, with the genome maintenance protein EBNA1 being expressed from an alternative promoter Qp, in addition to the BARTS and non-coding EBER transcripts (Figure 3 ). More recently, a subset of 3 from 20 endemic BL tumors have been identified that display a Wp-restricted form of EBV latency34 where, in addition to the BARTS and EBER transcripts, EBNAs1, 3A, 3B, 3C and -LP are expressed from the Wp promoter (Figure 3 ); these tumors are distinct in carrying a mutant form of the viral genome with a deletion of the EBNA2 gene, such that EBNA2 and the EBNA2-induced LMP proteins are not expressed. Importantly, this form of restricted viral latency also appears to be compatible with the c-myc–driven growth program. Most recently, we identified an endemic tumor which, though homogeneous for the t(8:14) translocation and other genetic changes, was nevertheless heterogeneous at the single-cell level with respect to EBV infection. This tumor, Awia-BL, yielded multiple subclones in vitro, some of which showed Latency I infection, others Wp-restricted latency, and others a novel “EBNA2+, LMP1–” latency characterized by expression of all six EBNAs again in the absence of LMPs (Figure 3 ); once again, all three types of subclone retained typically high c-myc levels and a BL-like cellular phenotype.35
The frequency of EBV’s association with BL (particularly endemic BL), and the fact that all EBV+ BLs (whether endemic, sporadic or AIDS-associated) carry the virus in every cell of the malignant clone, implies a definitive role for the virus in tumor pathogenesis, but the nature of that role has long remained in doubt. Hints that the virus might complement the deregulated c-myc gene by acting as an anti-apoptotic rather than as a growth-promoting agent first came from work with an EBV+ sporadic tumor, Akata-BL, which is unusual in that it generates rare EBV-genome–negative cells in late passage.36 Comparison of EBV+ and EBV– Akata-BL clones revealed that a Latency I EBV infection provided the cells with a slight, but significant, protection from apoptotic triggers. Similar observations have since been seen with rare endemic BL lines yielding EBV-loss subclones in late passage37 and now more importantly from an early passage tumor.35 The mechanism of protection mediated by Latency I infection, and indeed the viral gene product responsible, remains a contested issue.37,–41 By contrast, Wp-restricted latency was found to provide much stronger protection from apoptosis,35,42 perhaps explaining why this form of infection is represented relatively frequently in endemic BL (vis-à-vis Latency I) despite its establishment being dependent on the very rare circumstance of infection with an EBNA2 gene–deleted virus. Very interestingly, recent work has shown that Wp-restricted tumors are distinguished from Latency I BLs in showing greatly reduced expression of the pro-apoptotic Bcl2 family protein, Bim43 (Kelly et al, manuscript in preparation). This therefore provides a conceptual link between EBV’s postulated role as an anti-apoptotic agent in BL pathogenesis and the work in murine models identifying uncoupling of the c-myc/Bim connection as a key factor in lymphomagenesis driven by mutant c-myc alleles. It will be interesting to determine whether endemic BLs with Wp-restricted latency are more likely than Latency I tumors to carry a wild-type c-myc allele. This would be consistent with the hypothesis that one or more of the additional viral gene products expressed in Wp-restricted tumors actively downregulate Bim and could therefore substitute for the effects of c-myc mutation (see model postulated in Figure 2D ).
Holoendemic Malaria and HIV infection as Predisposing Factors in BL Pathogenesis
Often ignored in discussions of BL are the potentially important insights into tumor pathogenesis that could come from studying the other predisposing infections, namely (1) the widely accepted role of holoendemic Plasmodium falciparum malaria as the environmental factor underlying the 100-fold higher incidence of endemic BL (all EBV+) compared with that of the sporadic tumor in Western societies, and (2) the even more dramatic increase in BL incidence (both EBV+ and EBV– disease) seen in individuals infected by HIV.
It is firstly important to note that AIDS-BL is not a product of profound immune impairment but typically arises as an early symptom of AIDS at a time when patients still retain T-cell competence and display persistent lymphadenopathy rather than lymphodepletion. Furthermore, early work on HIV-associated lymphadopathy lesions clearly showed highly expanded germinal center activity,44 consistent with expansion of the very stage of B-cell differentiation where c-myc translocation and subsequent mutation would be most anticipated. The heightened risk of BL imposed by HIV is therefore most likely due to the still poorly understood ability of this virus to activate the B-cell system. Interestingly, the EBV load in circulating B cells is also elevated to a new steady-state level in the early years following primary HIV infection, and this again appears to be linked to direct effects upon the B-cell system and not to any detectable impairment of EBV-specific T-cell surveillance.45
Likewise, malaria is known to provide a chronic stimulus to the B-cell system, and several recent studies have shown that EBV loads are remarkably high in African children generally, and in children with malaria in particular.46,47 It remains to be seen to what extent holoendemic malaria’s role as a cofactor in BL development reflects its global B-cell stimulatory capacity per se and to what extent its gross disturbance of the EBV-host balance is mediated either as a byproduct of the above stimulation or by other mechanisms (reviewed in Rochford et al48). In this latter context, there is preliminary evidence of an inhibitory effect of acute malaria on EBV-specific T-cell surveillance.49 However, there is stronger evidence that the cysteine-rich interdomain region 1α (CIDR1α) of the malaria protein P falciparum erythrocyte membrane protein 1 (PfEMP1), expressed at the surface of parasitized red blood cells can activate B cells, in particular the memory B-cell pool in which EBV persists.50,51 This could have the dual effect of expanding the pool of latently infected cells in vivo and, by inducing the reactivation of some infected cells into EBV lytic cycle,52 of increasing total EBV loads. These various possibilities are not mutually exclusive. The interplay of malarial infection, EBV-host balance and BL risk therefore represents an important area for future work, with the potential to reveal new insights into BL pathogenesis.
Conclusions
Two seminal observations underpin the status of BL as a key model for the understanding of multistage tumorigenesis. First, the discovery in the 1960s of its association with EBV became a foundation stone of human tumor virology and has presaged four decades of research in which the full oncogenic potential of this virus has slowly been realised. Second, the discovery in the 1980s of c-myc/Ig gene fusions at the site of t(8:14), t(2:8) and t(8:22) translocations in BL united the worlds of cyto- and molecular genetics, setting a path that would open up the molecular basis of oncogenic change in many other tumor contexts. Ironically, so wide became the vistas in both the above fields as to divert attention away from BL itself. The present review shows how, more recently, that focus has been restored in a way that again highlights the continued potential of BL to reveal new insights. Thus, the analysis in transgenic mouse models of c-myc–driven B-cell lymphomagenesis is identifying key aspects of c-myc function that are relevant to BL pathogenesis, with a focus on the decoupling of c-myc’s proproliferative and pro-apoptotic effects. At the same time, work in the EBV field is now revealing the capacity of non–growth-transforming forms of EBV latency to confer protection from apoptosis. While not yet fully understood at a mechanistic level, these findings clearly point to ways in which EBV’s contribution to BL pathogenesis might be reconciled with the primacy of c-myc as the driver of tumor growth. Despite these advances, it is clear that many questions still remain to be resolved. These include questions about the timing of EBV’s contribution vis-à-vis that of the c-myc translocation, the influence of EBV on the molecular signature of the tumor, the identity of the BL progenitor cell and whether its germinal center–like phenotype truly reflects the tumor’s origin, and the mechanisms by which malaria and HIV increase BL risk. Addressing these issues will require the concerted efforts of many disciplines, the intelligent use of both in vivo and in vitro models, and a renewed focus on the analysis of primary tumor material from endemic, sporadic and AIDS-associated forms of the disease.
Three epidemiologically distinct forms of Burkitt lymphoma (BL).
| Form of BL . | Geographic location . | Incidence, per 100,000/y . | % EBV association . | Cofactors . |
|---|---|---|---|---|
| *Endemic BL (e-BL) occurs where Plasmodium falciparum malaria is holoendemic; incidence rates from African data. | ||||
| †Values for sporadic BL incidence and EBV association vary with location worldwide. In Northern Europe and the U.S., the disease occurs with 100-fold lower incidence than e-BL, and 15–20% tumors are EBV+. In some other areas (North Africa, North Brazil) incidence is only 10-fold lower than e-BL, and 85% of tumors are EBV+. | ||||
| ‡Values are based on data from untreated AIDS cohorts in the early years of the AIDS epidemic in the U.S. and Europe. | ||||
| Endemic* | Equatorial Africa and Papua New Guinea | 5-10, for age 3-12 y | 100 | Holoendemic malaria |
| Sporadic† | Worldwide, varies with location | 10- to100-fold less than e-BL | 15-85 | ? |
| AIDS-assoc‡ | Worldwide, data from untreated HIV carriers | 10- to 100-fold more than e-BL | 30-40 | HIV |
| Form of BL . | Geographic location . | Incidence, per 100,000/y . | % EBV association . | Cofactors . |
|---|---|---|---|---|
| *Endemic BL (e-BL) occurs where Plasmodium falciparum malaria is holoendemic; incidence rates from African data. | ||||
| †Values for sporadic BL incidence and EBV association vary with location worldwide. In Northern Europe and the U.S., the disease occurs with 100-fold lower incidence than e-BL, and 15–20% tumors are EBV+. In some other areas (North Africa, North Brazil) incidence is only 10-fold lower than e-BL, and 85% of tumors are EBV+. | ||||
| ‡Values are based on data from untreated AIDS cohorts in the early years of the AIDS epidemic in the U.S. and Europe. | ||||
| Endemic* | Equatorial Africa and Papua New Guinea | 5-10, for age 3-12 y | 100 | Holoendemic malaria |
| Sporadic† | Worldwide, varies with location | 10- to100-fold less than e-BL | 15-85 | ? |
| AIDS-assoc‡ | Worldwide, data from untreated HIV carriers | 10- to 100-fold more than e-BL | 30-40 | HIV |
Mechanisms for escaping c-MYC induced apoptosis. Panels A–C are adapted from Dang et al.28 (A) Acute activation of Myc induces target genes involved in proliferation, but the activation of ARF, p53, and Bim (which inhibits Bcl2) leads to apoptosis or cell cycle arrest. Activation of both the ARF/p53 and Bim pathways are required for apoptosis induction (B) Chronic expression of wild-type Myc induces lymphomagenesis coordinately with the inactivation of ARF or p53. (C) Chronic expression of Myc mutants derived from Burkitt lymphoma (BL) cells fail to activate Bim and hence promote lymphomagenesis despite the presence of wild-type p53 or ARF. (D) In our proposed model, EBV in a Wp-restricted form of latency down regulates Bim in BL cells and thus contributes to lymphomagenesis.
Mechanisms for escaping c-MYC induced apoptosis. Panels A–C are adapted from Dang et al.28 (A) Acute activation of Myc induces target genes involved in proliferation, but the activation of ARF, p53, and Bim (which inhibits Bcl2) leads to apoptosis or cell cycle arrest. Activation of both the ARF/p53 and Bim pathways are required for apoptosis induction (B) Chronic expression of wild-type Myc induces lymphomagenesis coordinately with the inactivation of ARF or p53. (C) Chronic expression of Myc mutants derived from Burkitt lymphoma (BL) cells fail to activate Bim and hence promote lymphomagenesis despite the presence of wild-type p53 or ARF. (D) In our proposed model, EBV in a Wp-restricted form of latency down regulates Bim in BL cells and thus contributes to lymphomagenesis.
Alternative forms of EBV latency. Transcripts expressed in lymphoblastoid cell lines (LCLs) and in three alternative forms of latency seen in early passage BL cell lines are illustrated relative to the EBV linear genome, with promoters (arrowheads) and splicing patterns as shown; coding exons (EBNA and LMP mRNAs) are shaded solid, while non-coding exons (EBER RNAs, BamHI A RNAs, and BART miRNAs) are unfilled.
Alternative forms of EBV latency. Transcripts expressed in lymphoblastoid cell lines (LCLs) and in three alternative forms of latency seen in early passage BL cell lines are illustrated relative to the EBV linear genome, with promoters (arrowheads) and splicing patterns as shown; coding exons (EBNA and LMP mRNAs) are shaded solid, while non-coding exons (EBER RNAs, BamHI A RNAs, and BART miRNAs) are unfilled.
Cancer Research UK Institute for Cancer Studies, The University of Birmingham, Birmingham, UK
Acknowledgments
The authors apologize to colleagues whose primary research papers are not cited because of the limited number of references. The authors’ studies are funded by Cancer Research UK.