Abstract
To reduce the risk of hemorrhage, experts advocate prescribing the anticipated therapeutic dose to patients who are beginning coumarin therapy, but until now there was no accurate way to estimate that dose. Using pharmacogenetics-based coumarin therapy, clinicians can now estimate the therapeutic dose by genotyping their patients for single nucleotide polymorphisms (SNPs) that affect coumarin metabolism or sensitivity.
SNPs in the cytochrome P450 complex (CYP2C9) affect coumarin metabolism. Patients with either of two common variants, CYP2C9*2 or CYP2C9*3, metabolize coumarins slowly and are twice as likely to have a laboratory or clinical adverse event, unless their initial coumarin doses are reduced. SNPs in vitamin K epoxide reductase (VKORC1) correlate with coumarin sensitivity. Patients known to be homozygous for a common VKORC1 promoter polymorphism, −1639 G>A (also designated as VKOR 3673, haplotype A, or haplotype*2), should be started on lower coumarin doses than genotype GG patients. By providing an estimate of the therapeutic coumarin dose, pharmacogenetics-based therapy may improve the safety and effectiveness of coumarin therapy.
Each year, millions of people take warfarin and other coumarins. Although these vitamin K antagonists are remarkably effective at preventing cardioembolic stroke, myocardial infarction, and venous thrombosis, they double the incidence of hemorrhage.1 The hemorrhage risk is greatest during the first weeks to months of therapy.2–6 To reduce this risk, experts advocate prescribing the anticipated therapeutic dose to patients who are beginning warfarin,7–9 but until now there was no accurate way to estimate that dose.10 By using pharmacogenetics-based warfarin therapy, clinicians can now estimate the therapeutic warfarin dose by genotyping their patients for single nucleotide polymorphisms (SNPs) that affect warfarin metabolism or sensitivity.
The objective of pharmacogenetics-based coumarin therapy is to improve the safety and the effectiveness of anticoagulant therapy. Recently, investigators and clinicians have used pharmacogenetics to develop dosing algorithms that estimate the warfarin dose based on genetic and clinical factors. In this paper we summarize the pharmacokinetics, pharmacodynamics, and pharmacogenetics that affect coumarin therapy.
Need for a Pharmacogenetics-Based Approach
Pharmacogenetics-based warfarin therapy should improve the safety and efficiency of warfarin initiation
Because warfarin is a dangerous drug that is commonly prescribed, improving its dosing could prevent hemorrhages. Already, over two million North Americans take warfarin therapy. This figure will rise as the North American population ages11 and atrial fibrillation (a disease of the elderly) becomes more prevalent. Pharmacogenetics-based therapy might reduce medical expenditure by preventing inpatients from being kept in the hospital until their therapeutic dose has been determined empirically.
Current approaches to warfarin induction fail to prevent adverse events
Prospective studies consistently identify warfarin induction, rather than warfarin maintenance, as the period when the INR is most likely to be out of range12,13 and when the rate of iatrogenic adverse events is greatest.2–5 For example, in the Columbus study of 1292 patients with an acute venous thromboembolism (VTE), both recurrent VTE and major bleeds were clustered in the first few weeks of therapy.4 Likewise, others have found that the INR is supra-therapeutic more than one-third of the time during the first month of usual care.5 In summary, standard warfarin induction is accompanied by a high rate of adverse events.
In an attempt to decrease the toxicity of warfarin induction, several dosing algorithms have been proposed,14–19 but none has been widely accepted. A major barrier to their implementation is that most were developed for middle-aged inpatients who could tolerate doses of 5–10 mg warfarin daily and who had daily monitoring of the international normalized ratio (INR).14,15 Today, the typical person taking warfarin is elderly.11,20,21 Because warfarin dose requirements decrease with advancing age,22 use of existing algorithms tends to overdose this growing population.16–19 Another problem is that the risk of overdose is increased now that most patients begin warfarin in the outpatient setting, where daily INR monitoring is not feasible (except with patient self-monitoring). The major flaw of existing warfarin algorithms is that they are empiric: they rely on trial-and-error dosing after an initial warfarin dose of 2 to 10 mg, rather than being tailored to individual genetic and clinical factors.23 As detailed below, researchers have developed warfarin-dosing algorithms that use clinical pharmacology, pharmacogenetics and clinical factors to estimate the warfarin dose. Clinicians will be able to use these algorithms to estimate the warfarin dose a priori, thereby decreasing the risk of overdose and hemorrhage during warfarin induction.
Coumarin Pharmacology
Pharmacology of warfarin
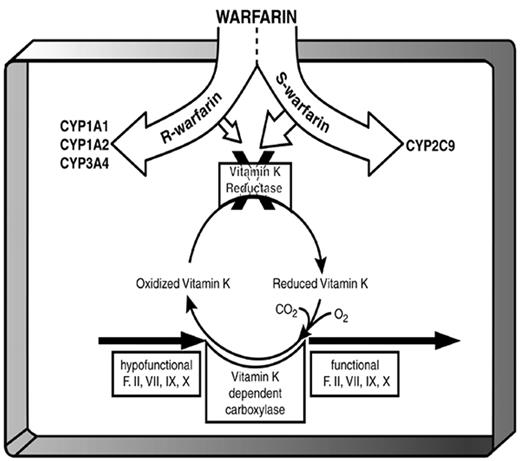
After oral administration, warfarin is completely absorbed and then 99% bound to albumin in the plasma. The free warfarin is taken up by the liver where it is biologically active and where it is metabolized by cytochrome P450 (CYP). Commercially available warfarin is a racemic mixture. The S-enantiomer is converted to 6- and 7-hydroxy-warfarin by CYP2C9 and eventually excreted in the bile, while the R enantiomer is metabolized by CYP1A1, CYP1A2, and CYP3A4 to an inactive alcohol that is excreted in the urine.24 The S-enantiomer more strongly blocks the regeneration of the reduced form of vitamin K, thereby interfering with the vitamin-K–dependent carboxylation of glutamic acid residues on coagulation factors prothrombin (II), VII, IX, and X and other vitamin-K dependent proteins (e.g., proteins C, S, and Z and osteocalcin). The pharmacologic result is that warfarin inhibits the formation of functional clotting factors.
In patients who are homozygous for the wild-type allele (CYP2C9*1), S-warfarin is cleared normally, resulting in a modest elevation of the INR. In contrast, patients with either of two SNPs in this gene have impaired metabolism of S-warfarin, the more active enantiomer of warfarin (Figure 1 ). The SNP in exon 3 (CGT>TGT) is denoted as CYP2C9*2, while the SNP in exon 7 (ATT>CTT) is called CYP2C9*3. Patients with one or two of these SNPs have reduced warfarin requirements and a 2–3 fold elevated risk of an adverse event when beginning warfarin.13,25,26 Also in exon 7 is the rarer CYP2C9*5 variant whose effect on warfarin metabolism in vivo is unknown.27
Pharmacology of other coumarins resemble pharmacology of warfarin
Like S-warfarin, S-acenocoumarol inhibits vitamin K epoxide reductase and is metabolized by CYP2C9. An important pharmacokinetic difference is that S- and R-warfarin have half lives of approximately 32 and 43 hours (respectively) while S- and R-acenocoumarol have half lives of 2 and 8 hours (respectively). Because of the very short half life of S-acenocoumarol, R-acenocoumarol is the more important enantiomer. However, genetic polymorphisms that affect warfarin pharmacology also affect acenocoumarol pharmacology.28,29
Like S-warfarin and S-acenocoumarol, both S- and R-phenprocoumon inhibit vitamin K epoxide reductase and are partly metabolized by CYP2C9.30 (S- and R-phenprocoumon are also partly metabolized by CYP3A4.30) Phenprocoumon has a much longer half life than either acenocoumarol or warfarin: about 172 hours for the more potent S-phenprocoumon and 156 hours for R-phenprocoumon.31 Genetic polymorphisms that affect warfarin and acenocoumarol pharmacology also affect phenprocoumon.29
Pharmacogenetic Determinants of Warfarin Metabolism
In vivo, two CYP2C9 SNPs have been associated with increased responsiveness to warfarin.13,32–36 Aithal and colleagues compared controls who required typical warfarin doses to patients whose therapeutic warfarin dose was ≤ 10.5 mg/wk. Patients requiring low doses were more likely to have a supratherapeutic INR at the time of warfarin induction, almost 4 times more likely to bleed, and 6 times more likely to have the CYP2C9*2 or CYP2C9*3 SNPs.13,36 Others found that CYP2C9*3 decreased selectivity of CYP2C9 for S-warfarin and that residue 359 was a component of the warfarin binding site.37,38
Other CYP2C9 SNPs have less clinical relevance
In 2001, while genotyping 32 Japanese patients who were slow metabolizers of phenytoin, Imai and colleagues found a new polymorphism designated as CYP2C9*4: a T1076C transversion led to an Ile359Thr substitution.43 This mutation, named 2C9*4, was not present in 100 unselected Japanese volunteers or in 369 Americans.44 Later, it was determined that this SNP was in CYP2C19, rather than CYP2C9, as originally reported.45
In 2002, Dickmann and colleagues found a C1080G transversion that leads to an Asp360Glu substitution.46 They found this mutation, CYP2C9*5, in 4 of 120 African-American participants and in 0 of 140 European-American participants. The clinical relevance of this SNP is not known. However, because it is a non-synonymous SNP that is adjacent to the CYP2C9*3 SNP, it might impair the metabolism of S-coumarins.
Kidd and colleagues reported a null polymorphism, 818delA, which they named CYP2C9*6.47 The patient presented with an overdose of phenytoin (an anticonvulsant that is metabolized by CYP2C9) and was found to have a phenytoin clearance that was only 17% of normal. In summary, warfarin metabolism is impaired in individuals who are heterozygous or homozygous for CYP2C9*2 or CYP2C9*3 (Table 1 ), but other CYP2C9 SNPs require further validation before being used to tailor the warfarin dose.
Clinical and laboratory outcomes with CYP2C9 SNPs
The CYP2C9 SNPs are associated with a 2- to 3-fold increased risk of bleeding during warfarin induction,13,25,26,48 but not during long-term therapy.40 This observation suggests that pharmacogenetics-based coumarin therapy will affect the initial warfarin dose(s), but will be less important once the therapeutic dose is known.
Pharmacogenetic Determinants of Warfarin Sensitivity
Coumarins such as warfarin inhibit the action of vitamin K epoxide reductase, whose gene (VKORC1) has been recently discovered by two independent teams.49,50 In the absence of coumarin, the vitamin K cycle regenerates reduced vitamin K1 from its epoxide (Figure 1 ). Reduced vitamin K is a cofactor for post-translational γ-carboxylation of glutamic acid residues on several proteins, including coagulation factors II (prothrombin), VII, IX, and X. Although coumarins also inhibit the γ-carboxylation of anticoagulant proteins C, S, and Z, inhibition of clotting factor activity is their main pharmacologic effect. γ-Carboxylation allows for normal hemostasis by resulting in negatively charged γ-carboxyglutamates on factors II, VII, IX, and X, which bind to calcium cations and then to platelet phospholipid membranes. γ-Carboxylation is also required for the development of other tissues, and warfarin exposure in utero can cause mental retardation, nasal hypoplasia, and limb or digit abnormalities in the fetus.51
Identification of the VKORC1 gene
By the same mechanism, coumarins are effective rodenticides, causing fatal bleeding in coumarin-naïve rodents. Studies of wild-caught, coumarin-resistant rats have provided important clues to the location of VKOR. For example, Kohn and Pelz52 used linkage analysis to place Rw, the gene for warfarin resistance in the rat, near one of several microsatellite markers. Then, they identified three candidate human chromosome locations for Rw, one of which was on the short arm of chromosome 16.
The second clue to the location of VKORC1 came from a study of familial multiple coagulation factor deficiency (FMFD). FMFD is an extremely rare, autosomal recessive, bleeding disorder characterized by inadequate γ-carboxylation of coagulation factors II, VII, IX, and X. Fregin and colleagues investigated two kindreds with FMFD with suspected VKORC1 complex mutations due to biochemical evidence of elevated vitamin K epoxide/vitamin K hydroquinone ratios.53 Genome-wide scanning identified a marker on chromosome 16 with a LOD score of approximately 3, and haplotype analysis confirmed homozygosity for chromosome region 16p12–q21 in one of the families. Mapping studies of Rw in the rat and a similar gene in mice (war) identified a homologous region on human chromosome 16, and several known genes that flank Rw and War are located on the short arm of chromosome.16,52,54
In 2004, Rost, Fregin and colleagues identified VKORC1 in this region and confirmed its function by transfecting the novel gene into a cell line.50 To identify the gene, they directly sequenced genomic DNA in a 4-Mb region on chromosome 16 from 2 probands with FMFD and 4 unrelated patients with warfarin resistance. They found a 5126 base-pair gene (GenBank id # gi:13124769; IMAGE 3455200) of 3 exons coding a 163-amino acid protein. They named this gene vitamin K epoxide reductase complex subunit 1 (VKORC1). Northern blotting showed high expression in liver and heart cells. Each resistant patient had unique missense SNPs in the new gene, while the FMFD patients were homozygous for an identical C292T missense SNP that predicted an Arg98Trp substitution. Next, they sequenced 26 coumarin-resistant rats and found an A416C missense mutation in all of them; 15 coumarin-sensitive rats all lacked this SNP. They confirmed VKORC1 function by transfecting human embryonic kidney cells with the novel gene. The gene from the coumarin-resistant rat (Rw), conferred VKORC1 activity that was minimally inhibited by warfarin.
In the same issue of Nature, Li and colleagues used positional cloning to identify VKORC1 in the chromosome region 16p12–q21.49 They focused on 13 candidate genes of unknown function with sequence motifs predictive of transmembrane proteins. By using short-interfering RNA (siRNA) they were able to degrade each gene sequentially in a lung carcinoma cell line. They found that MGC11276 mRNA, which mapped to 16p11.2, was required for VKOR activity. They confirmed the activity of VKORC1 by transfecting insect cells (S frugiperda) with the gene and demonstrating that the cells gained warfarin-sensitive VKORC1 activity. In summary, elegant experiments found a novel reductase with VKOR activity. In vivo, the reductase likely functions with other proteins to form a complex in the endoplasmic reticulum.
SNPs in VKORC1 correlate with warfarin dose
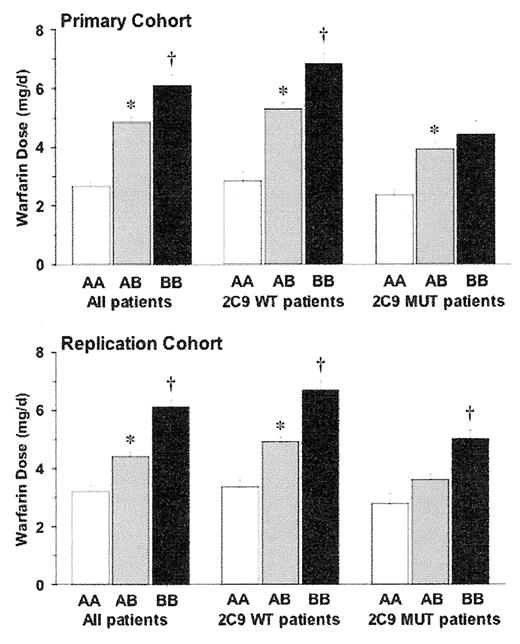
Investigators at the University of Washington (Seattle) and Washington University (St. Louis) recently identified informative SNPs in VKORC1.55 They performed direct resequencing of PCR amplicons encompassing the upstream promoter region, intragenic sequence, and the downstream region of VKORC1. They found 10 (all non-coding) common SNPs and defined a set of 4 SNPs that could be used to infer haplotype. A single promoter SNP, designated 6853 G>C [rs17886369], predicted 21%–25% of the variability in the warfarin dose in Caucasian patients taking warfarin. The mean therapeutic doses of warfarin per day differed significantly based on this SNP (and its corresponding haplotype): 2.7–3.4 mg for genotype CC (haplotype AA), 4.3–4.9 mg for genotype C/G (haplotype A/B), and 6.0–6.2 day for genotype GG (haplotype BB) (P < 0.001) (Figure 2 ). The prevalence of these genotypes differed by race and contributed to lower warfarin doses in Asian and Mexican populations and the greater doses in African ancestry populations.55,56 The 6853 SNP correlated with greater gene expression of vitamin K epoxide reductase activity suggesting that carriers of the G allele require greater warfarin doses because they have greater endogenous epoxide activity.
Italian investigators sequenced VKORC1 in 147 patients taking warfarin therapy and discovered 4 SNPs. The 1173 C>T [rs9934438] SNP in intron 1 correlated with the therapeutic warfarin dose, predicting 14% of the variability in the warfarin dose. A Dutch study also found that patients with the 1173 C>T SNP had a higher risk of bleeding when beginning phenprocoumon,29 but this relationship was not significant in new users of acenocoumarol. (The Dutch investigators did not examine patients beginning warfarin.)
Taiwanese investigators sequenced VKORC1 in 11 warfarin-sensitive patients (maintenance warfarin dose ≤ 1.5 mg/day) and 5 warfarin-resistant patients (maintenance warfarin dose ≥ 6 mg/day). They identified a promoter polymorphism, −1639 G>A (also designated as VKOR 3673 or rs9923231) that was in high linkage disequilibrium with the VKORC1 1173 C>T polymorphism and haplotype A, using the nomenclature of Rieder et al,55 and correlated with less VKORC1 expression in a transfected cell line. All warfarin-resistant patients carried the G allele at −1639 while none of the warfarin-sensitive patients did. In 104 randomly selected Han Chinese patients taking warfarin, only 21 patients carried this allele and their average warfarin dose was 1 mg/day (38%) greater than 83 patients who lacked this polymorphism.
Swedish investigators genotyped 98 warfarin-treated patients for common SNPs in VKORC1.57 The found that patients with VKORC1*2 haplotype (similar to haplotype A, using the nomenclature of Rieder et al55) had lower warfarin doses and lower therapeutic warfarin levels than other patients. Furthermore, these patients also had a higher percentage of INR values outside the therapeutic interval.
Pharmacogenetic-based Dosing Algorithms
Another group of Swedish investigators genotyped 201 Caucasian patients for common SNPs in VKORC1.58 They found a VKORC1 non-coding SNP [rs2359612] in intron 2 that was linked with the above non-coding SNPs and explained 29% of the variability in warfarin dose. They combined this SNP, the CYP2C9*2 and CYP2C9*3 SNPs, and clinical factors (weight, age, interacting drugs, and indication for warfarin treatment) to derive a regression model that accounted for 56% of the variability in the warfarin dose.
British researchers developed a dosing algorithm based on 297 patients on a stable warfarin dose.59 All patients were Caucasian and had a target INR of 2–3. The equation that best estimated the warfarin dose (mg/day) was: [0.628 − 0.0135 × age − 0.24 × CYP2C9*2 − 0.37 × CYP2C9*3 −0.241 × VKORC1 + 0.0162 × height (in cm)]2, where CYP2C9 genotype is 0, 1, or 2 for the number of *2 and *3 alleles within the patient’s genotype and VKORC1 − 1639 genotype is 1 for GG, 2 for GA, and 3 for AA. They found an R2 of 54% in the derivation cohort and retrospectively validated the algorithm in 38 patients.
More recently, we collected DNA, demographic variables, laboratory values, and medication histories from 1015 participants taking warfarin. After PCR amplification, we genotyped for three coding CYP2C9 SNPs, five VKORC1 SNPs, and one factor II (F2) SNP. A VKORC1 promoter SNP (3673 G>A also known as −1639 G>A) was the first variable to enter the stepwise regression equation and was associated with a 28% [95% CI: (25% to 30%)] decrease in the warfarin dose per allele. Other key predictors of dose were: body surface area (+11% per 0.25 m2), CYP2C9*3 (−33% per allele), CYP2C9*2 (−19% per allele), and age (−7% per decade). A pharmacogenetic equation that included these factors, race, smoking status, amiodarone use, target INR, and indication for warfarin therapy, explained 53% of the variability in the warfarin dose. The dosing algorithm is available on a free website: www.WarfarinDosing.org.
In summary, investigators in three continents have confirmed that common SNPs in VKORC1 correlate with the therapeutic warfarin dose. Although estimates vary, either of the two VKORC1 SNPs (6853 or −1639) explain about one-fourth of the variability in the therapeutic warfarin dose. By combining a VKORC1 SNP with CYP2C9 SNPs and clinical factors, pharmacogenetics can estimate a therapeutic coumarin dose, potentially improving the time in the target INR range in coumarin-naïve patients. Because of this potential benefit, several companies are developing rapid genotyping assays for CYP2C9 and VKORC1 based on microchips, bead technology, or fluoresceinated DNA probes to identify biotinylated amplicons. The FDA is considering revising the package insert for warfarin therapy to accommodate pharmacogenetics-based therapy and they may sponsor a clinical trial to quantify the safety and effectiveness of this approach. One unstudied issue that the proposed trial could address is whether genotype could be used to optimize dose refinements, just as it could be used to select initial coumarin dose.
Although CYP2C9 and VKORC1 are the current genes that could be used in a trial or clinical care, future genetic targets that may strengthen pharmacogenetics approach. 165Thr>Met of the factor II (F2) gene, −402G>A of the factor VII (F7) gene, and (CAA repeat) of the γ-glutamyl carboxylase gene predicted warfarin sensitivity in a Japanese population60 but require validation in other populations. Two rare mutations in the propeptide of factor IX also lead to warfarin sensitivity61,62 but may be too rare to be included in commercial genotyping platforms. A SNP in calumenin (CALU) was associated with warfarin sensitivity in a single patient,63 and direct sequencing of this gene may yield clinically relevant SNPs. In another study, a SNP in exon 26 of the adenosine triphosphate-binding cassette gene ABCB1 (multidrug resistance gene 1) was over-represented among low-dose patients,64 but this SNP also awaits validation.
Cytochrome P450 2C9 single nucleotide polymorphisms (SNPs) that are known to affect warfarin metabolism.
| . | Prevalence (%) . | . | . | ||||
|---|---|---|---|---|---|---|---|
| Designation . | Asian . | White . | Black . | Protein Change . | SNP . | Effect on Warfarin Dose . | Sample References . |
| † CYP2C9*1 is the wild-type allele. | |||||||
| CYP2C9*1 † | 98.2 | 80.3 | 94.2 | none | none | referent | |
| CYP2C9*2 | 0 | 12.7 | 3.4 | Arg144Cys | C430T | −14% to −20% | 26,39–42 |
| CYP2C9*3 | 1.8 | 7.0 | 1.5 | Ile359Leu | A1061C | −21% to −49% | 26,39–42 |
| . | Prevalence (%) . | . | . | ||||
|---|---|---|---|---|---|---|---|
| Designation . | Asian . | White . | Black . | Protein Change . | SNP . | Effect on Warfarin Dose . | Sample References . |
| † CYP2C9*1 is the wild-type allele. | |||||||
| CYP2C9*1 † | 98.2 | 80.3 | 94.2 | none | none | referent | |
| CYP2C9*2 | 0 | 12.7 | 3.4 | Arg144Cys | C430T | −14% to −20% | 26,39–42 |
| CYP2C9*3 | 1.8 | 7.0 | 1.5 | Ile359Leu | A1061C | −21% to −49% | 26,39–42 |
After oral absorption, warfarin is transported to the liver where CYP1A1, CYP1A2, and CYP3A4 metabolize the R-enantiomer and CYP2C9 metabolizes the more potent S-enantiomer. Warfarin inhibits vitamin K reductase, which is synthesized (at least in part) byVKORC1. By impairing the regeneration of the reduced form of vitamin K, R- and S-warfarin interfere with the vitamin-K–dependent carboxylation of clotting factors prothrombin (II), VII, IX, and X.
Reprinted with permission from Gage BF, van Walraven C, Pearce L, et al. Selecting patients with atrial fibrillation for anticoagulation: stroke risk stratification in patients taking aspirin. Circulation. 2004;110:2287–2292.
After oral absorption, warfarin is transported to the liver where CYP1A1, CYP1A2, and CYP3A4 metabolize the R-enantiomer and CYP2C9 metabolizes the more potent S-enantiomer. Warfarin inhibits vitamin K reductase, which is synthesized (at least in part) byVKORC1. By impairing the regeneration of the reduced form of vitamin K, R- and S-warfarin interfere with the vitamin-K–dependent carboxylation of clotting factors prothrombin (II), VII, IX, and X.
Reprinted with permission from Gage BF, van Walraven C, Pearce L, et al. Selecting patients with atrial fibrillation for anticoagulation: stroke risk stratification in patients taking aspirin. Circulation. 2004;110:2287–2292.
Effect of VKORC1 haplotype on warfarin dose stratified by VKORC1 haplotype and CYP2C9 status: either wild type (WT) or CYP2C9*2 and/or CYP2C9*3 mutants (MUT).
Primary cohort: UW (N = 185); Replication cohort: Wash U (N = 368).
Reprinted with permission from Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293.
Effect of VKORC1 haplotype on warfarin dose stratified by VKORC1 haplotype and CYP2C9 status: either wild type (WT) or CYP2C9*2 and/or CYP2C9*3 mutants (MUT).
Primary cohort: UW (N = 185); Replication cohort: Wash U (N = 368).
Reprinted with permission from Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293.
Acknowledgments: The author would like to acknowledge the assistance of Charles Eby, MD.