Abstract
Classical Hodgkin lymphoma (HL) is characterized by the presence of Reed-Sternberg (RS) cells, which are transformed post-germinal center B cells destined for apoptosis since they have not undergone successful immunoglobulin gene rearrangement. Several mechanisms, including latent infection by Epstein-Barr virus (EBV), allow these cells to survive. It is remarkable that many of the signaling pathways that promote survival are shared between the EBV-induced proteins, such as EBNA1, LMP1, and LMP2, and other molecules that are upregulated in RS cells. A key role is played by the presence of constitutive nuclear factor (NF)-κB, which is induced by LMP1, as well as by CD30, CD40, tumor necrosis factor (TNF)-α, and Notch1 interactions, and results in the upregulation of at least 45 genes including chemokines, cytokines, receptors, apoptotic regulators, intracellular signaling molecules, and transcription factors. The other characteristic of classical HL is the presence of an extensive inflammatory infiltrate. Key features of this infiltrate are that it comprises Th2 and T regulatory cells and generally lacks Th1 cells, CD8 cytotoxic T cells, and natural killer (NK) cells. The RS cells appear to induce this infiltrate by the secretion of Th2 type chemokines such as TARC and MDC. The RS cells also produce cytokines that inhibit Th1 responses, as interleukin (IL)-10 and transforming growth factor (TGF)-β express CD95 ligand, which induces apoptosis of activated Th1 and CD8 T cells. Other important mechanisms that allow the RS cells to escape an effective anti-EBV immune response include the downregulation of HLA class I in EBV-negative cases or the presence of a polymorphism in HLA class I in EBV-positive cases that allow escape from CD8-mediated cytotoxicity. On the other hand, expression of HLA-G allows the escape from NK cells that would normally recognize the HLA class I-negative RS cells. Overall, the cellular infiltrate in HL appears to play a decisive role in allowing the RS cells to survive by providing an environment that suppresses cytotoxic immune responses and providing cellular interactions and cytokines that support the growth and survival of RS cells. Future therapeutic strategies could focus directly on the NF-κB activation, on various receptors to ligand interactions, on the chemokine and cytokine network, or on the induction of effective anti-EBV latent protein immune responses.
Hodgkin lymphoma (HL) is a B cell lymphoma. More precisely, Hodgkin lymphoma comprises at least two B cell lymphoma entities. What these lymphomas have in common is the presence of so-called Reed-Sternberg (RS) cells and an extensive immune response. In the case of nodular lymphocyte predominance type Hodgkin lymphoma (NLP HL), the evidence for the B cell nature is overwhelming and the L&H type RS cells can be considered neoplastic germinal center cells. This includes expression of BCL-6 and frequent translocations or rearrangements involving the bcl-6 gene.1 The nature of the RS cells in classical HL (cHL) is much more ambiguous: genotypically they generally have immunoglobulin gene rearrangements, but phenotypically they lack most B cell characteristics and frequently express genes normally expressed in T cells, macrophages, and dendritic cells. When compared to normal B cells, classical RS cells resemble activated, post-germinal center B cells. This includes expression of a number of genes normally expressed in plasma cells, such as CD138 and MUM1/IRF4.2
Role of Epstein-Barr Virus
With the emerging view that classical RS cells derive from post-germinal center B cells, destined for apoptosis in the B cell selection process because of the lack of successful immunoglobulin gene rearrangement, the question arises: What enables these precursors to escape apoptosis? For many years a role of a virus, specifically Epstein-Barr virus (EBV), was suspected based on epidemiological and serological findings. The first patient with EBV-positive RS cells was described 20 years ago.3 Since then, it has become clear that in the Western world 20%–40% and in Asia and South America more than 70% of HL cases are EBV positive.4 In EBV-positive HL, latent infection of a B lymphocyte by the virus is now accepted to be an early transforming event. Although EBV normally persists as a harmless passenger and adapts to normal B-cell developmental pathways, the virus can also have a marked influence on differentiation processes of B cells. In particular, EBV can replace the survival signals that are normally provided by the B-cell receptor (BCR) during germinal center reaction, thereby circumventing the normal apoptotic pathway and allowing the survival of BCR-deficient B cells.5 If EBV provides an important part of the answer to the rescue of RS cell precursors from apoptosis, then what is the mechanism in EBV-negative cases? As yet, the question of whether EBV might operate through a “hit and run” tactic in these cases or whether another virus is involved is unanswered.
Latency Type II in cHL
A limited number of EBV-induced molecules have been identified in classical EBV-positive RS cells. This so-called latency type II pattern includes EBNA1, latent membrane protein-1 (LMP1) and LMP2, and two small RNAs, termed EBER 1 and 2. Expression of EBNA1 is maintained in all EBV latency states and all EBV-associated tumors, most likely because of its critical functions. The EBNA1 protein initiates EBV episome replication before mitosis, and it anchors the viral episome to mitotic chromosomes during cell division. This ensures persistence of EBV DNA in proliferating cells and, therefore, maintains EBV-encoded small RNA genes, which contribute to oncogenicity in BL.6 It is of interest that in vitro infection of the HL cell line KMH2 with EBV leads to expression of EBNA-1, and that CD40 ligand and IL-4 can induce LMP-1, but not EBNA-2.7
Several EBV-encoded proteins mimic key signaling pathways in B cells: LMP1 shares several features with CD40, a member of the tumor necrosis factor receptor (TNFR) family 1. LMP1 and CD40 both activate the transcription factor nuclear factor-κB (NF-κB) by promoting turnover of IκBα, an important inhibitor of NF-κB.8 CD40 associates with CD40L that is expressed, for example, on the surface of the T lymphocytes surrounding RS cells, whereas LMP-1 signals independently of a ligand and is dependent on self-association. Activated CD40 and LMP-1 co-localize in lipid rafts and recruit TRAF3, JAK3, and/or TRADD.9
Key Role of Constitutive Nuclear NF-κB in Reed-Sternberg Cells
What are the functional consequences of constitutive nuclear NF-κB in H/RS cells? NF-κB, which was originally discovered in the nucleus of B cells as a factor binding to the kappa light chain of immunoglobulin, has emerged as a central player in the transformation of RS cells. NF-κB is present in its inactive state in the cytoplasm of almost every cell type, but when activated, it translocates to the nucleus, binds DNA, and regulates the expression of over 200 different genes. Overexpression of a dominant negative version of IκBα in several Hodgkin cell lines resulted in decreased nuclear NF-κB, decreased proliferation rates, an enhanced apoptotic response to removal of serum from the culture medium, and decreased ability to form tumors in immunodeficient mice.10 In a molecular profiling study of two HL cell lines, 45 genes were identified whose expression was regulated by NF-κB. These included chemokines, cytokines, receptors, apoptotic regulators, intra-cellular signaling molecules, and transcription factors.11
In addition to LMP1 and CD40, two other TNF family receptor–ligand pairs have the potential to activate NF-κB in HL: TNF-α produced by the RS cell may act in an autocrine fashion via TNF-α receptors,12 and CD30 on the H/RS cells may be cross-linked by CD30 ligand on either the H/RS cell or on surrounding cells.13 Finally, NF-κB activity is also regulated via transcriptional control by Notch-1.14 Notch1 belongs to a family of transmembrane receptors that control cell proliferation and differentiation in response to extracellular ligands expressed on neighboring cells. During lymphoid development, Notch1 plays a critical role in the T-cell/B-cell lineage decision. Notch pathway activation induces translocation of intracellular Notch (ICN) to the nucleus, where it interacts with the transcription factor CSL (CBF1/RBP-Jk, Suppressor of Hairless, Lag-1). In vitro, ICN binds Mastermind-like proteins (MAML), which act as potent Notch coactivators. Notch1 is highly expressed on RS cells. Interaction between intact Notch1 and its ligand Jagged1 dramatically induces proliferation and inhibition of apoptosis in vitro. In HL, Jagged1 is expressed in RS cells as well as in bystander cells (endothelial cells, smooth muscle cells, and epithelioid histiocytes). Notch1 signaling may therefore be activated in tumor cells by Jagged1 through homotypic or heterotypic cell-cell interactions with neighboring RS or bystander cells.15 Some of the key interactions involved in NF-κB activation in RS cells are summarized in Table 1 .
Function of Notch1 Signaling
Notch-1 signals in a manner similar to EBNA2, which is not expressed in latency state II. Although not related by sequence, the cellular Notch protein and EBNA2 share several biochemical and functional properties, such as interaction with CBF1 and the ability to activate transcription of a number of cellular and viral genes.16 Both Notch and EBNA2 activate an important subset of cellular genes associated with type III latency and B-cell growth, while EBNA2 more efficiently induces important viral genes, such as LMP-1. EBNA-2 and Notch both upregulate CD21 and downregulate immunoglobulin μ (Igμ) expression. Activation of the Notch-related signaling pathways may represent a key mechanism by which EBNA2 contributes to EBV-induced immortalization.17 The effect of Notch signaling can be mediated by the early transcription factor GATA-2. GATA-2 expression was found in RS cells in 50% of cHL cases, while nodular lymphocyte-predominant HL, Burkitt lymphoma, and diffuse large B-cell lymphoma did not express GATA-2, and normal germinal center cells were also negative. The function of GATA-2 is to maintain hematopoietic stem and early progenitor cells in an undifferentiated state. Thus GATA-2 expression may cause RS cells to re-acquire certain aspects of undifferentiated progenitor cells.18
Resistance to Death Receptor-Mediated Apoptosis
RS cells express CD95 on their membranes in the vast majority of cases, but resist CD95-induced apoptosis. Interaction of CD95 with its ligand CD95L normally results in a rapid formation of the death-inducing signaling complex (DISC) containing Fas-associated death domain-containing protein (FADD), caspase-8, and caspase-10. FADD overexpression is frequently detectable in HL. Mutations of the FAS gene as found in the autoimmune lymphoproliferative syndrome (ALPS) are associated with a 7-fold increased risk of developing HL. However, only 5% of HL cases not associated with ALPS are associated with FAS mutations.19 Cellular FADD-like interleukin 1β-converting enzyme-inhibitory protein (c-FLIP) inhibits the apoptotic pathway by binding to the DISC. c-FLIP is expressed in a majority of cases of cHL.20,21 By confocal microscopy c-FLIP was shown to be predominantly localized at the cell membrane of RS cells. Despite expression of other NF-κB-dependent antiapoptotic proteins, the selective down-regulation of c-FLIP by small interfering RNA oligoribonucleotides was sufficient to sensitize HRS cells to CD95 and TNF-related apoptosis-inducing ligand-induced apoptosis. Therefore, c-FLIP is a key regulator of death receptor resistance in HRS cells.21
LMP2A Mimics the B Cell Receptor (BCR)
LMP2A is capable of providing B cells with survival signals in the absence of normal B-cell receptor (BCR) signaling and blocks normal BCR-signaling in vitro. Furthermore, expression of LMP2A in developing B cells in vivo induces a global downregulation of genes necessary for normal B-cell development. LMP2A increases the expression of genes associated with cell cycle induction and inhibition of apoptosis, alters the expression of genes involved in DNA and RNA metabolism, and decreases the expression of B-cell-specific factors and genes associated with immunity. Many alterations in gene expression induced by LMP2A are similar to those in RS cells, suggesting that LMP2A expression in EBV-infected B cells may lead to the induction and maintenance of an activated, proliferative state that could ultimately result in the development of HL.22
Immune Response Dominated by RegulatoryT Cells
As EBV latent infection of B lymphocytes is very common, usually occurs early in life, and is persistent, it can be speculated that immune responses are capable of repressing early or precursor stages of HL. Indeed, immunodeficiency syndromes are associated with increased HL incidence. Human immunodeficiency virus (HIV)-seropositive individuals are 3 to 7 times more likely to develop HL, in particular EBV-positive HL.23 Infectious mononucleosis (IM) is associated with an increased risk of developing EBV-positive HL, especially in young adults, but also of developing EBV-negative HL.24 Another question is what causes the prominent, but apparently ineffective, immune response in HL. Lymph nodes involved by HL generally contain only a minority of RS cells suspended in an abundant inflammatory infiltrate, suggesting that immunological mechanisms contribute to HL pathogenesis. The inflammatory infiltrate consists of predominantly CD4+ T cells, with a variable admixture of CD8+ T cells, B cells, plasma cells, eosinophils, histiocytes, neutrophils, and mast cells and usually comprises more than 99% of the tumor mass. Some of the interactions between RS cells and other cell types are illustrated in Figure 1 (see Color Figures, page 548). The abundance of inflammatory cells may reflect the fact that RS cells produce and secrete large amounts of chemokines (in particular TARC and MDC) that attract cells expressing the CCR4 receptor, such as activated Th2 lymphocytes.25 The various chemokines secreted by RS cells are summarized in Table 2 . RS cells also secrete a number of cytokines, including interleukin (IL)-5, -6, -9, -10, and -13.26 In particular, the cytokine IL-13 and also its receptor IL-13Rα1 were found to be highly expressed in Hodgkin cell lines and RS cells.27 This important cytokine promotes T helper cell differentiation to the Th2 phenotype and can indirectly promote immunosuppression and influence the survival and/or proliferation of B cells by binding to IL-13Rα1 and activating STAT6.28 The explanation for such a wide variety of cytokine expression is unknown, but in a recent study we found expression of several transcription factors, including T-bet and GATA-3, that direct Th1 and Th2 type cytokine production, respectively, by T cells.29
The cytokines, chemokines, and various other receptors expressed in HL are summarized in Figure 2 (see Color Figures, page 548). Another question is why the immune response is not effective against the tumor cells. HL infiltrating lymphocytes are anergic to stimulation with many mitogens and also suppress peripheral blood mononuclear cell (PBMC) responses.30 This appears to be caused by IL-10–secreting T cells as well as CD4+CD25+ regulatory T cells. The immunosuppressive effect of the HL infiltrating cells can be neutralized with anti-IL-10, by preventing cell-to-cell contact, and with anti CTLA-4. The lymphocytes in HL do not produce cytokines, such as IL-2, IL-4, and inter-feron (IFN)-γ with primary (KLH) and recall (PPD) antigens and the mitogen ConA. However, when stimulated with PHA or with phorbolester (PMA)-ionomycin, they are capable of producing these cytokines. When the CD26– CD4 cells immediately surrounding the Reed-Sternberg cells are purified and stimulated with PMA ionomycin, they produce IL-4 and IFN-γ. The potential to produce IL-4 was the reason these cells were previously considered Th2 like. Absence of IL-2 production upon stimulation is also associated with anergy. The exact nomenclature of this cell population is thus a matter of semantics.31 In addition to the IL-10–producing cells (Tr1) and CD4+CD25+ regulatory cells, there are also transforming growth factor (TGF)-β–producing cells (Th3) present in the infiltrate. There are variations in the lymphocyte populations involved in different cases.
It can be concluded that as an overall population the infiltrating lymphocytes do not have Th1 type functions and are probably attracted into the tissues by chemokines TARC and MDC as CCR4-expressing Th2 cells. Although these cells do not spontaneously produce IL-2 or IL-4, they produce IL-10 despite not being fully activated and therefore function as Tr1 cells. The major remaining question is what causes the predominance of T cells with suppressor activity in HL. It appears that RS cells, although they have the genotype of B cells, execute a functional program with expression of molecules like CD40, CD80, and CD86 that is similar to that of antigen-presenting cells, but results in tolerance. Mechanisms include the production of immunosuppressive cytokines like IL-10, especially in EBV-positive cases, or IL-13 and TGF-β, especially in nodular sclerosis cases.26 A number of strategies have been proposed for HRS cells to escape cytotoxic killing. HRS cells express Fasligand (CD95L) and RCAS1 (receptor binding cancer antigen expressed on SiSo cells) cell surface proteins that can induce apoptosis in NK cells and cytotoxic T lymphocytes (CTLs).32,33 Proteinase inhibitor 9 (PI9), a serpin that inhibits granzyme B, may also protect HRS cells from immune system-mediated apoptosis.34 Finally, the RS cells themselves are protected by overexpression of cFLIP or infrequently by FAS mutation.19
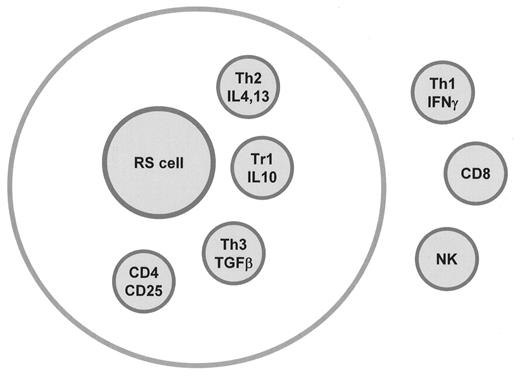
The relevance of these findings is that they may allow a better design of new treatment modalities. There are indications that the infiltrating cells in fact support the growth and survival of the RS cells. Therefore, blocking chemokines such as TARC and MDC to prevent the influx of T cells may be effective. Alternatively, interference with binding or signaling of IL-13 or other cytokines might also be of potential therapeutic value. On the other hand, blocking of the immunosuppressive signals, such as provided by IL-10 and TGF-β or the removal of the suppressor regulatory T cells, may enhance cytotoxic T-cell responses. The T cell subsets present in cHL are illustrated in Figure 3F1 .
T Cell Immunity Directed Against EBV Antigens
The RS cells of EBV-positive HD express several tumor antigens derived from the virus and could, in principle, be a target for adoptive immunotherapy with viral antigen-specific T cells. It is remarkable that HRS cells have generally lost the majority of B cell markers (CD19, CD20, CD22, and CD79a), but consistently express molecules involved in antigen presentation (HLA, CD74, TAP1, and TAP2), costimulation (CD40, CD70, CD80, and CD86), and cell adhesion (ICAM-1 and LFA-3).35 Despite the consistent deregulated B cell phenotype, HRS cells maintain their professional antigen-presenting phenotype. Antigenic peptides that potentially can be presented by HRS cells may be derived from proteins that are altered by malignant transformation. In addition, in EBV-positive HL, antigenic peptides may be derived from LMP1, LMP2, and EBNA1. Presentation of these antigenic peptides is not strictly HLA class I dependent, as transformation-related antigenic peptides in general and EBNA1 antigenic peptides in particular are at least partially HLA class II restricted.36,37 However, in EBV-positive HL only sporadic lymphocytes specific for EBV latent antigens have been found in the involved tissues.38 Several mechanisms enable the RS cells to escape a CD8-mediated cytotoxic or an NK cell response. Like most tumor-associated antigens in immunocompetent hosts, the potential target antigens are only weakly immunogenic, consisting of LMP1 and LMP2 antigens and EBNA-1. CD8+ T cells specific for LMP1 are infrequent and for LMP2 are observed only in HLA-A2-positive individuals. One might then hypothesize that HLA-A2-negative individuals would be more susceptible to EBV-positive HL than HLA-A2-positive individuals. However, the frequency of HLA-A2 in both EBV-positive and EBV-negative HL patients is the same as in the general population.39,40 In addition, RS cells may escape immune control by MHC class I restricted CD8+ T cells in several ways. First, the tumor cells have down-regulated the expression of most latent antigens, except LMP1, LMP2, and EBNA1. Second, EBNA1 contains a Gly/Ala domain that prevents cytosolic degradation by inhibiting proteasomal digestion, which is necessary for HLA class I-mediated presentation.41 This disables CD8 T cell-mediated cytotoxicity as one of the two main CTL effector functions of T cell-mediated cytotoxicity. On the other hand, CD4 T cells may play a role that enables most healthy carriers of EBV to avoid latency I and II malignancies.42 In mouse models, these T cells are important for resistance to cancer and viruses, including virally induced malignancy. Activated CD4+ T cells deliver signals to dendritic cells, which, in turn, are vital to the induction and expansion of antigen-specific CD8+ T lymphocytes. Since the CD8+ T-cell arm of the EBNA1 response is blocked at the level of antigen presentation on MHC class I, the CD4+ response becomes the principal T-cell mechanism to recognize EBNA1. Because in normal carriers the EBNA1 response is of the Th1 type that conveys resistance to viruses and tumors, this protein provides a new focus for vaccination and immunotherapy.43 Reciprocally, a loss of the Th1 phenotype of the EBNA1 response, for instance by a shift to a Th2 or a T regulatory response, may allow for the development of EBV-associated malignancies. As indicated above, several mechanisms, including chemokines and cytokines like IL-10, IL-13, and TGF-β, can skew the CD4 response against EBNA1, LMP1, and LMP2 to the Th2, Th3, or Tr1 type, thereby reducing T cell-mediated resistance. Treatment strategies employing adoptive transfer of cytotoxic T cells should take the above mechanisms into account.44 The various T cell subtypes involved in T cell immunity for EBV are summarized in Table 3 .
Role of HLA in Susceptibility for HL
Genetic predisposition observed in HL seems related to immune system function, as a large number of HLA associations with HL have been reported and may partially account for the observed genetic susceptibility to HL. However, there is little consensus on the role of specific HLA alleles and haplotypes. As EBV antigenic peptides can be presented to the immune system by HLA class I and II, and selection of these peptides depends on HLA alleles, HLA class I and II associations are both expected to be important in EBV-positive HL. On the other hand, HLA class II associations may be related to attraction and activation of inflammatory cells and may influence HL subtype. In addition, non-HLA genes located in the HLA class III region and dispersed between the HLA genes in the class I and II subregions may be involved in overall HL pathogenesis. Genetic associations of all three HLA subregions with HL susceptibility have been reported; however, these associations may not pinpoint causative genetic variations because of the strong linkage disequilibrium within the HLA region.45 Further genetic screening is necessary to identify the exact HL susceptibility loci and to determine whether antigen presentation is important in the pathogenesis of HL.
Two different mechanisms may explain HLA associations in HL. The first one involves antigen presentation by HLA class II to CD4+ T helper lymphocytes in the surrounding infiltrate. Lymphocytes in the inflammatory component are essential in HL pathogenesis, and it is possible that presentation of antigenic peptides by HLA is important in (the early) attraction and activation of these lymphocytes. RS cells in addition need immunomodulatory properties to overcome the induction of an effective immune response. Because HLA class II is expressed in virtually all cases of HL this mechanism may be important in overall HL pathogenesis. In addition, presentation of different antigenic peptides may induce different inflammatory reactions leading to the observed spectrum of HL subtypes.
The second mechanism involves HLA class I restricted antigen presentation to CD8+ cytotoxic T lymphocytes. RS cells in a high percentage of EBV-negative HL do not express HLA class I, whereas RS cells in most EBV-positive cases do show HLA class I cell surface expression.46.47 In the latter group HRS cells should be able to present EBV-derived antigenic peptides leading to the induction of EBV-specific cytotoxic responses. Since it is known that subdominant immunogenic targets LMP1 and LMP2 are restricted to common HLA haplotypes, it can be hypothesized that less common HLA class I haplotypes prohibit efficient immune responses and thereby predispose to the development of EBV-positive HL.
In a retrospective, population-based study, patients with HL were reclassified according to the World Health Organization classification and EBV status was assessed by in-situ hybridization of EBV-encoded small RNAs. Germline DNA was isolated from 200 patients diagnosed between 1987 and 2000 and from their first-degree relatives. Genotyping was done with 33 microsatellite markers spanning the entire HLA region and two single nucleotide polymorphisms in the genes for TNF α and β. Classic association analysis and the haplotype sharing statistic were used to compare patients with controls. Classic association analysis (but not the haplotype sharing statistic) showed an association of consecutive markers D6S265 and D6S510 (P = 0.0002 and 0.0003), located in the HLA class I region, with EBV-positive patients. The haplotype sharing statistic (but not classic association analysis) showed a significant difference in mean haplotype sharing between patients and controls surrounding marker D6S273 (P = 0.00003), located in HLA class III. Areas within the HLA class I and class III regions are associated with susceptibility to HL, the association with class I being specific for EBV-positive disease.48 This finding strongly suggests that antigenic presentation of EBV-derived peptides is involved in the pathogenesis of EBV-involved HL. Polymorphisms in the HLA region could explain ethnic variation in the incidence of HL. The association of EBV-positive HL with HLA class I suggests that this polymorphism might affect the proper presentation of EBV antigens to cytotoxic T lymphocytes.48
The HLA-G protein has been implicated in evasion of CTL- and NK cell-mediated cytotoxicity. In EBV-negative cHL cases expression of MHC class I is frequently lost, providing escape from CTL lysis, but potentially inducing NK cell responses. We therefore investigated expression of HLA-G in cHL and correlated this with EBV status and expression of MHC class I (HLA-B/C and β2-microglobulin). We found expression of HLA-G by HRS cells in 67% of cHL cases, which was associated with absence of MHC class I (P = 0.005) and EBV negative status (P = 0.005). This expression could be attributed to the soluble HLA-G5 isoform, as shown by Western blotting and reverse-transcription polymerase chain reaction (RT-PCR) analysis.49 Secretion of soluble HLA-G by HRS cells may therefore contribute to escape from immunosurveillance in classical Hodgkin lymphoma.
Conclusions
Classical HL is characterized by the presence of RS cells, which are transformed post-germinal center B cells destined for apoptosis since they have not undergone successful immunoglobulin gene rearrangement. Several mechanisms, including latent infection by EBV, allow these cells to survive. It is remarkable that many of the signaling pathways that promote survival are shared between the EBV-induced proteins, such as EBNA1, LMP1, and LMP2, and other molecules that are upregulated in RS cells. A key role is played by the presence of constitutive nuclear NF-κB, which is induced by LMP1, as well as by CD30, CD40, TNF-α, and Notch1 interactions, and results in the upregulation of at least 45 genes including chemokines, cytokines, receptors, apoptotic regulators, intracellular signaling molecules, and transcription factors. The other characteristic of cHL is the presence of an extensive inflammatory infiltrate. Key features of this infiltrate are that it comprises Th2 and T regulatory cells and generally lacks Th1 cells, CD8 cytotoxic T cells, and NK cells. The RS cells appear to induce this infiltrate by the secretion of Th2 type chemokines like TARC and MDC. The RS cells also produce cytokines that inhibit Th1 responses, as IL-10 and TGF-β express CD95 ligand, which induces apoptosis of activated Th1 and CD8 T cells. Other important mechanisms that allow the RS cells to escape an effective anti-EBV immune response include the downregulation of HLA class I in EBV-negative cases, or the presence of a polymorphism in HLA class I in EBV-positive cases that allow escape from CD8-mediated cytotoxicity. On the other hand, expression of HLA-G allows the escape from NK cells that would normally recognize the HLA class I-negative RS cells. Overall, the cellular infiltrate in HL appears to play a decisive role in allowing the RS cells to survive by providing an environment that suppresses cytotoxic immune responses and providing cellular interactions and cytokines that support the growth and survival of RS cells. Future therapeutic strategies could focus directly on the NF-κB activation, on various receptors to ligand interactions, on the chemokine and cytokine network, or on the induction of effective anti-EBV latent protein immune responses.
NF-κB activation.
| Abbreviations: NF, nuclear factor; LMP, latent membrane protein; TNF, tumor necrosis factor. |
|
| Abbreviations: NF, nuclear factor; LMP, latent membrane protein; TNF, tumor necrosis factor. |
|
Chemokines expressed in Reed-Sternberg cells.
|
|
T cell immunity for Epstein-Barr virus.
| • However, only few cells present and several mechanisms that would skew repertoire toward Th2, Th3 or Tr. |
|
| • However, only few cells present and several mechanisms that would skew repertoire toward Th2, Th3 or Tr. |
|
T-cell subsets in Hodgkin lymphoma.
Abbreviations: RS, Reed-Sternberg; TGF, transforming growth factor.
T-cell subsets in Hodgkin lymphoma.
Abbreviations: RS, Reed-Sternberg; TGF, transforming growth factor.