Abstract
Hematological complications occur frequently in patients with both primary and secondary immunodeficiency disorders. Anemia, thrombocytopenia or leukopenias may bring these individuals to the attention of hematologists. Conversely, evidence suggesting a lymphoproliferative disorder may be the cause for referral. This session will provide an update on the diagnosis and treatment of immunodeficiency diseases ranging from isolated defects in antibody production to the severe combined immunodeficiencies (SCID).
Immunodeficiency diseases have traditionally been defined as defects in the development and function of T and B cells, the primary effector cells of specific cellular and humoral immunity. However, it has become increasingly evident that innate immune mechanisms contribute greatly to host defense, either through acting alone or by enhancing specific T and B cell responses.
In Section I, Dr. Lewis Lanier reviews the burgeoning information on the extensive families of activating and inhibitory immunoreceptors that are expressed on NK cells, dendritic cells, T and B cells, and phagocytic cells. He provides an overview on the biological functions of these receptors in host defense.
In Section II, Dr. Mary Ellen Conley defines the spectrum of antibody deficiency disorders, the most frequently occurring types of primary immunodeficiencies. She covers the different defects in B-cell development and function that lead to antibody deficiencies, and includes diagnosis and therapy of these disorders.
In Section III, Dr. Jennifer Puck discusses the diagnosis and treatment of the different types of SCID. She describes the genetic basis for SCID, and the benefits, pitfalls, and complications of gene therapy and bone marrow transplantation in SCID patients.
I. KIR, LIR, Fc Receptors and Related Leukocyte Receptors and Their Biological Functions
Lewis L. Lanier, PhD*
Department of Microbiology and Immunology and the Cancer Research Institute, University of California San Francisco, 513 Parnassus Avenue, Box 0414, San Francisco, CA 94143-0414
LLL is an American Cancer Society Research Professor and is supported by NIH grants CA89189, CA89294 and CA095137.
The function of the immune system is to detect and eliminate pathogens and possibly transformed cells. T cells and B cells are endowed with cell surface receptors generated by genetic recombination and the introduction of non-germline encoded nucleotides that provide for limitless diversity and the potential to interact with essentially any foreign substance. These receptors provide for antigen-specific recognition, the hallmark of an adaptive immune response. By contrast, most hematopoietic cells (e.g., monocytes, macrophages, granulocytes, dendritic cells and natural killer [NK] cells) do not have receptors generated by genetic recombination. Certain receptors of the innate immune system (e.g., toll-like receptors and scavenger receptors, reviewed in Janeway and Medzhitov1) operate by detecting pathogen-encoded molecules that are conserved between different organisms (for example, certain carbohydrates or glycolipids unique to bacteria, double-stranded RNA encoded by viruses, etc.). An alternative strategy is provided by the existence of receptors (e.g., NKG2D2) that recognize host-encoded “stress-induced” proteins that are not present at high levels in healthy tissues but are induced by infection or transformation. These stimulatory immune receptors that activate cells in the presence of perceived danger are complemented by a system of inhibitory receptors that are designed to prevent or dampen immune responses in the absence of pathogenic insult, thereby avoiding autoimmunity. Many of the inhibitory receptors recognize cell surface glycoproteins that are constitutively expressed on healthy cells, for example major histocompatibility complex (MHC) class I. Inhibitory receptors are found not only on cells of the innate immune system, but are also present on T and B lymphocytes and can regulate responses initiated through their antigen-specific receptors. The vigor of an immune cell’s response is determined by the intracellular integration of signals transmitted by the activating and inhibitory receptors. This review will focus on selected families of receptors that regulate immune responses and will present examples of their potential roles in certain human diseases.
Biochemical Basis for Immune Activation Versus Inhibition
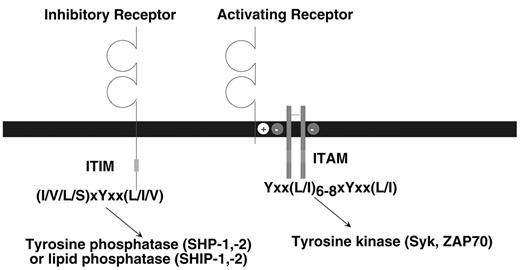
The class of stimulatory immune receptors described in this review share a common biochemical pathway for cellular activation. As depicted in Figure 1 , the ligand-binding and signaling functions are mediated by different protein subunits within the receptor complex (reviewed in Malissen3). The ligand-binding receptor lacks intrinsic signaling activity; a small adapter protein provides this activity. The ligand-binding receptor and signaling adapter assemble by interactions between their transmembrane domains. Typically, a positively charged amino acid (i.e., K or R) in the receptor forms a salt bridge with a negatively charged amino acid (i.e., D or E) in the adapter. The adapter proteins signal by virtue of one or more immunoreceptor tyrosine-based activation motifs (ITAM) (YxxL/I x6–8YxxL/I where x denotes any amino acid with 6 to 8 amino acids between the two YxxL/I elements) in their cytoplasmic domains. When the receptor engages ligand, clustering of the receptor complex results in phosphorylation of the tyrosines in the ITAM, which is likely mediated by a Src-family tyrosine kinase. The phosphorylated ITAM then serves as a docking site for either the Syk or ZAP70 tyrosine kinase, triggering a downstream cascade of activation events that can lead to cell-mediated cytotoxicity, cytokine production and proliferation. In addition to the ITAM-bearing adapter proteins that have been described in the B and T cell antigen receptor complexes, ITAM are also present in the FcεRIγ, ζ and DAP12 adapter proteins that are used by many different immune receptors in myeloid and NK cells.
The inhibitory receptors are all characterized by one or more immunoreceptor tyrosine-based inhibition motifs (ITIM), (I/V/L/S)xYxx(L/I/V) where x denotes any amino acid) in the cytoplasmic domains (reviewed in Ravetch and Lanier4). Binding of ligand to the extracellular domain causes, by an unknown mechanism, tyrosine phosphorylation of the ITIM, probably by a Src-family kinase. In turn, the phosphorylated ITIM may bind to two classes of SH2-containing inhibitory signaling effector molecules, the inositol phosphatases SHIP-1 or SHIP-2 (Src homology 2 domain-containing inositol-5′ phosphatase) or the tyrosine phosphatases SHP-1 or SHP-2 (Src homology 2-containing phosphatase). It appears that different ITIM-bearing receptors show a preference for association with SHP-1, SHP-2, SHIP-1, or SHIP-2 based on in vitro assays; however, a precise structural basis for this selectivity has not been defined. In general, SHP-1 and SHIP appear to be the dominant players, depending on the particular receptor studied. Moreover, there is no clear consensus concerning the critical substrates of the tyrosine phosphatases that inhibit cellular activation. This may depend upon the activation pathway being affected and the cell type involved. There is a growing family of receptors in B cells, T cells, NK cells and myeloid cells that express ITIM and serve to regulate the activation of these cells.4 In mice, loss-of-function mutations in these ITIM-bearing receptors can cause hyper-activation or autoimmunity.
Killer Cell Immunoglobulin-like Receptors
Killer cell immunoglobulin-like receptors (KIR; also designated CD158, see http://www.ncbi.nlm.nih.gov/prow/guide/679664748_g.htm) are encoded by a family of highly related genes that arose by gene duplication on human chromosome 19q13.4.5 The number of KIR genes varies between different individuals; presently, there are at least 15 functional genes and two pseudogenes (for an updated list see www.ebi.ac.uk/ipd/kir/)6 (Table 1 ). Not all genes are present in an individual; rather several haplotypes with different numbers of KIR genes have been defined that are represented within the population. In addition, there is allelic polymorphism that adds further diversification. KIR are transmembrane-anchored type I glycoproteins that possess either two or three Ig-like domains in the extracellular region (referred to as KIR2D or KIR3D, respectively). The inhibitory KIR have long cytoplasmic domains with ITIM (designated KIR2DL or KIR3DL). The activating KIR have short cytoplasmic domains lacking signaling capacity (KIR2DS or KIR3DS); these associate with DAP12 to provide signaling function.7 KIR2DL4 is unusual in that it has a functional ITIM and also a charged amino acid in the transmembrane, but does not associate with DAP12. This KIR apparently can trigger cytokine production, but not cytotoxicity.8
Only NK cells and a subset of memory T cells express KIR. The inhibitory KIR2DL molecules recognize polymorphic determinants of HLA-C and inhibit the effector functions of NK cells against cells expressing normal levels of MHC class I. Similarly, the inhibitory KIR3DL molecules recognize polymorphic HLA-B or HLA-A ligands. As yet, the physiological ligands of the activating KIR2DS and KIR3DS glycoproteins are not defined, although certain KIR2DS demonstrate weak binding to HLA-C. A remarkable feature is that an individual NK or T cell clone expresses only a subset of the KIR genes and the expression of each allele at a locus is controlled independently.5 This provides for extensive diversity with respect to the array of receptor combinations expressed within the total NK cell population. The KIR repertoire in an individual is stable over time and is largely dictated by the genotype, with subtle modifications imposed by the person’s MHC class I genotype, as determined by analysis of families in which KIR and HLA haplotypes segregate together or independently in the progeny.9 Each NK cell clone appears to express at least one inhibitory KIR that recognizes a self-MHC class I ligand, presumably to prevent auto-aggression; however, many NK cells express KIR that recognize products of MHC class I alleles not present in the individual.10 Based on a comparison of humans and chimpanzees, the KIR genes are diversifying and evolving at a rapid pace, particularly the genes encoding the activating receptors.11 This implies selective pressure, possibly by pathogens. This concept is supported by the recent observation that an activating NK receptor in mice (the Ly49H receptor that is functionally similar to the activating KIR in humans) recognizes a viral glycoprotein encoded by cytomegalovirus (CMV). Ly49H provides protection to CMV in strains of mice that express this activating receptor.12
The loss of MHC class I on a cell, due to viral infection or transformation, would be predicted to lower the threshold for attack by NK cells or memory T cells bearing an inhibitory KIR. Conversely, the presence of an activating KIR might provide positive recognition for cells expressing ligands for these receptors. The ability of inhibitory KIR to dampen or prevent human NK cell or T cell stimulation has been documented extensively by using in vitro experimental models. Emerging studies suggest a protective or detrimental role for KIR in certain human diseases. For example, based on epidemiological evidence Carrington and colleagues13 have shown a benefit in HIV-infected individuals who have the activating KIR3DS1 and an HLA-Bw4 allotype. In this case, an activating KIR may provide protection against progression towards AIDS in virus-infected individuals. By contrast, susceptibility to psoriatic arthritis has been correlated with having KIR2DS1 and/or KIR2DS2 genes, but only when HLA-C ligands for the inhibitory receptors, KIR2DL1 and KIR2DL2/3, are absent in the individual.14 Similarly, an unusual subset of CD4+ T cells, lacking CD28, from patients with rheumatoid vasculitis have been shown to express the activating KIR2DS2 and cross-linking this receptor stimulated T cell cytokine production, despite the fact these cells lacked the ITAM-bearing DAP12 adapter subunit.15 Another very exciting clinical observation suggests that hematopoietic stem cell transplants into recipients with an HLA-C haplotype that does not provide ligands for the donor’s inhibitory KIR may result in the development of donor-derived NK cells that attack the recipient’s residual acute myelogenous leukemia (AML), but do not cause graft-versus-host disease16 (reviewed in Parham and McQueen17). By contrast, other studies have not observed a benefit by mismatching KIR ligands in hematopoietic stem cell transplants.18,19 While these clinical experiments involve only small numbers of patients and no mechanisms have been defined to demonstrate a direct role for KIR in the disease, they are intriguing and provide an impetus for more extensive study.
Leukocyte Immunoglobulin-like Receptors (LIR)
The KIR gene cluster is located just telomeric on chromosome 19q13.4 of another related family of genes encoding activating and inhibitory immune receptors, designated LIR, MIR, ILT, CD85, or LILR (reviewed in Trousdale et al20 and Borges and Cosman21). The thirteen LIR genes segregate into two clusters surrounding the LAIR-1 and LAIR-2 genes (Table 1 ). The inhibitory LAIR-1 has one extracellular Ig-like domain and ITIM sequences in the cytoplasmic domain, whereas different members of the LIR family may have two or four Ig-like extracellular domains. Moreover, some LIR genes encode inhibitory receptors with two, three or four ITIM in their cytoplasmic domains, while other genes encode soluble LIR (e.g., LIR4) or LIR without intrinsic signaling capacity that associate with the ITAM-bearing FcεRIγ adapter and are activating receptors.22 Although adjacent in the genome to the KIR genes, the LIR and LAIR genes are considerably older and demonstrate much less allelic polymorphism (reviewed in Voltz et al23). Moreover, with exception of the absence of the LIR-4 (ILT-6) in some people, all individuals have the same composition of LIR genes. Thus, the LIR and LAIR genes are under different selective pressures than the KIR genes.
LIR are expressed abundantly on myeloid cells and B cells, as well as on some NK cells and T cells. Ligands have only been identified for two of LIR; LIR-1 (ILT2) and LIR-2 (ILT4) bind to MHC class I. In contrast to KIR, LIR-1 and LIR-2 recognize monomorphic residues in the conserved α3 domain of MHC class I.24 Therefore, LIR-1 and LIR-2 bind to essentially all human MHC class I proteins, including HLA-A, -B, -C, -E, -F, and -G. This may have provided the ancestral system for leukocytes to be inhibited when encountering other cells that express high levels of MHC class I. The KIR genes may have evolved to provide a more sophisticated surveillance mechanism whereby these receptors recognize individual alleles of a single MHC class I locus, permitting detection of aberrant cells that have lost only one MHC class I gene product, rather than all class I.
As yet, there is little knowledge about the role of LIR in immune functions in vivo. An intriguing hint comes from the fact that LIR-1 was discovered because of its ability to bind with high affinity (nM) to UL18, an MHC class I-like protein encoded by human CMV.25 Presumably, CMV may have evolved UL18 to suppress immune response to the virus by engaging an inhibitory receptor on leukocytes. A recent study investigating lung transplantation has noted a correlation between CMV infection and an increase in the percentage of the patient’s peripheral blood T cells expressing the inhibitory LIR-1.26 The number of patients studied was small and other viral infections were not analyzed as control; nonetheless, this is consistent with the concept that UL18 may function as a virulence factor by engaging an inhibitory receptor. A mouse CMV viral glycoprotein with homology to MHC class I has been shown to bind to an inhibitory Ly49I receptor present on NK cells in certain CMV-susceptible strains of mice, suggesting that a similar strategy has been used by this pathogen.12 Although not proven, it is tempting to speculate that LAIR may serve to dampen the activation of leukocytes in the intestine, thereby avoiding autoimmunity in this tissue.
Fc Receptors
Receptors on leukocytes that bind to the Fc region of immunoglobulins provide a critical link between humoral and cellular immunity. The importance of this system is underscored by the wealth of receptors devoted to this task (Table 2 ). A human Fc receptor for IgA (FcαR, CD89) is encoded by a gene telomeric to the KIR genes on chromosome 19q13.4 (Table 1 ). Myeloid cells express CD89, allowing them to phagocytose IgA-coated bacteria or yeast. Other Fc receptors bind IgA, including the polymeric Ig receptor and the Fcα/μ receptor; both also bind IgM (reviewed in Monteiro et al27). Whereas the predominant role of the polymeric Ig receptor is for transport of IgM and IgA across epithelial cell barriers in mucosal tissues, the Fcα/μ receptor is expressed on B cells and myeloid cells and appears to be involved in antigen uptake.
There are numerous Fc receptors that bind to IgG with differing affinities (reviewed in Ravetch and Bolland28). Some are activating receptors (e.g., CD16a, CD64) and use the ITAM-bearing FcεRIγ adapter protein that is shared with many other activating receptors complexes, including the high affinity IgE receptor from which its name was derived. Unlike other activating receptors, CD32a (FcγRIIa) has an ITAM in its cytoplasmic domain, providing for ligand recognition and stimulatory signaling by the same polypeptide. Upon binding to IgG-coated cells or immune complexes, myeloid cells or NK cells expressing activating IgG Fc receptors are triggered to secrete cytokines and to kill IgG-coated cells by a process known as antibody-dependent cellular cytotoxicity (ADCC). Unlike all other Fc receptors, the CD32b1 (FcγRIIB) receptor, which is expressed on B cells, has an ITIM in its cytoplasmic domain and functions as a negative regulator to inhibit signaling through the B cell receptor when the cells are exposed to immune complexes.29 This is accomplished by the recruitment of the inositol 5′-phosphatase SHIP to the phosphorylated tyrosine in the ITIM after engagement of the receptor by IgG. The FcRn receptor for IgG differs from other Fc receptors in that it has structural homology to MHC class I. The receptor was first identified on epithelial cells in the intestine of neonates, where it serves to capture IgG from maternal milk and transport it into the circulation of the neonate; thereby, providing passive immunity to the offspring.
FcεRI consists of an IgE-binding α chain, associated with two signaling proteins, FcεRIβ and FcεRIγ, both of which have ITAM. Unlike other Fc receptors, FcεRI binds with high affinity to monomeric IgE, thereby concentrating IgE, which is present in very low concentrations in the serum, onto the membrane of mast cells and basophils. IgE plays an important role in host defense against certain pathogenic parasites, but is detrimental in that it often causes fatal allergic responses against otherwise non-harmful antigens. CD23, a lectin-like receptor with low affinity for IgE, is present on B cells and appears to have a regulatory function by suppressing further production of IgE; however, the mechanism involved in the inhibition has not been defined.
Collectively, a major role of Fc receptors in the immune system is to impart antigen-specific recognition to cells of the innate immune system. In this manner, the effector functions of macrophages, neutrophils and NK cells can be targeted specifically against Ig-coated cells or pathogens. This can be beneficial in host defense against pathogens and can be exploited by the use of passive antibody therapy to treat infectious disease or cancer. However, the activating Fc receptors can also be detrimental in the presence of autoantibodies and immune complexes and can cause allergy and asthma (reviewed in Ravetch and Bolland28).
Related Immune Receptors
While originally appreciated in Fc receptors, it is now clear that many families of immune receptors possess related isoforms that have activating and inhibitory function (reviewed in Ravetch and Lanier4 and Lanier30). While beyond the scope of this article to comprehensively review this field, a few general principles are emerging. First, it seems likely that the inhibitory receptors function as a fail-safe mechanism to ensure that the immune response is tightly regulated to avoid autoimmune reactions toward healthy tissues. Loss-of-function genetic mutations in certain inhibitory receptors results in a hyper-reactive immune response and in some cases severe autoimmune disease. The role of the activating receptors in these paired inhibitory-activating receptor families has been more enigmatic. In many cases where ligands have been found for the inhibitory isoforms within a receptor family, the activating receptors have been shown not to recognize the same ligand as the inhibitory receptor, even when the activating and inhibitory isoforms differ by only a few amino acids in their extracellular domains. For example, in the KIR family the inhibitory receptors recognize MHC class I but the activating isoforms don’t bind or bind only with very low affinity. Similarly, two inhibitory receptors in the LIR family bind MHC class I, but MHC does not serve as a ligand for any of the activating LIR receptors. Although speculative, it is possible that ligands for the activating receptors are encoded by pathogens; one example of a viral glycoprotein functioning as the ligand for an activating Ly49 family NK cell receptor has been discussed previously. An alternative possibility is that ligands for the activating receptors will be encoded by host genes that are typically silent or expressed at only minimal levels until induced by infection with a pathogenic organism. Determining the biological role of these intriguing families of immune receptors will provide a fruitful field for investigation in the near future.
II. Defects in B Cell Development and Antibody Deficiency
Mary Ellen Conley, MD*
Department of Immunology, St. Jude Children’s Research Hospital, Memphis, TN and
Department of Pediatrics, University of Tennessee College of Medicine, Memphis, TN
University of Tennessee College of Medicine, St. Jude Children’s Research Hospital, 332 North Lauderdale, Memphis, TN 38105
Antibody deficiencies are a heterogeneous group of disorders that include both primary and secondary forms of the disease. Some of the primary forms are due to single gene defects1–,3 but others, such as common variable immunodeficiency, are due to complex combinations of genetic and environmental factors.4 Secondary forms of antibody deficiency include situations of decreased antibody production, as in patients treated with chemotherapy, and conditions associated with protein loss, such as the nephrotic syndrome. Regardless of the cause, antibody deficiencies result in an increased susceptibility to infection with encapsulated bacteria, particularly S. pneumoniae and H. influenzae. Patients with antibody deficiencies develop recurrent and/or persistent infections typical of these organisms such as pneumonia, otitis and sinusitis.4– 8 More severe and invasive infections caused by these organisms (for example: sepsis, meningitis, epiglottitis, cellulitis, empyema and septic arthritis) are also common. Neutropenia may be the presenting finding in patients with primary antibody deficiencies; therefore, it is critical that hematologists are familiar with these disorders.
Patients with primary antibody deficiencies should be treated with gammaglobulin replacement and aggressive use of antibiotics. We generally prescribe chronic prophylactic antibiotics for these patients. Referral to a center with expertise in immunodeficiency should be part of the management. There is excellent evidence that patients undergoing complex surgeries do better at institutions that perform many procedures; patients with hemophilia and sickle cell disease have better outcomes if they are treated at specialized centers. It is reasonable to assume that patients with rare immunodeficiencies will have fewer complications if they are cared for by physicians with experience caring for similar patients. In this review I will focus on patients with single gene defects resulting in absent or impaired antibody production.
Defects in Early B Cell Development
Defects in early B cell development are associated with the onset of recurrent bacterial infections in the first few years of life, profound hypogammaglobulinemia and markedly reduced or absent B cells in the peripheral circulation.5,6,9 An important physical clue leading to the diagnosis of B cell deficiency is the marked paucity or absence of tonsils and lymph nodes in affected patients. The bulk of lymph nodes primarily consists of germinal follicles, which are largely composed of B cells. In the absence of B cells, the germinal follicles do not develop and the nodes are very small. The majority of patients with early defects in B cell development, approximately 85%, have mutations in Btk, the gene responsible for X-linked agammaglobulinemia (XLA; also called Bruton’s agammaglobulinemia)1 (Table 3).2,10 Approximately 5% have mutations in μ heavy chain, the primordial immunoglobulin gene.9,11,12 A smaller number have defects in other components of the pre-B cell receptor, the surrogate light chain or the Igα/Igβ transmembrane signal transduction module.13–,15 Mutations in the scaffold protein BLNK also result in a severe defect in B cell development.16 Disorders associated with myelodysplasia can sometimes masquerade as B cell–specific disorders.17 In about 5% of patients with defects in early B cell development, the nature of the underlying abnormality remains unknown.
X-Linked Agammaglobulinemia
The identification of Btk as the gene responsible for XLA in 1993 was a major breakthrough in our understanding of normal B cell development.1,2 Btk is a hematopoietic specific cytoplasmic tyrosine kinase that is expressed in all blood cells except T cells and terminally differentiated plasma cells.18,19 It is a 659 amino acid protein containing a C terminal catalytic domain preceded by an SH2 domain, an SH3 domain, a proline rich region and an N terminal pleckstrin homology domain (PH domain). These interaction domains allow Btk to bind to upstream and downstream components of signal transduction pathways. Btk is activated by a variety of different receptors including the high affinity IgE receptor on mast cells,20 the glycoprotein VI collagen receptor on platelets,21 and the antigen receptor on the B cells.22,23 Each of these receptors uses a shared signal transduction pathway that is initiated by binding of a ligand to a transmembrane receptor containing an ITAM motif, a conserved cytoplasmic sequence with two tyrosines that act as docking sites when phosphorylated. A tyrosine kinase containing two SH2 domains, either Syk or Zap-70, is activated by binding to the ITAM motif. In turn Syk/Zap-70 phosphorylates a scaffold protein, either BLNK or SLP-76, depending on the cell line.24,25 Btk and PLCγ then assemble on the scaffold protein, allowing Btk to phosphorylate and fully activate PLCγ.26 Activated PLCγ cleaves phosphatidylinositol 4,5-bisphosphate to generate diacylglycerol, which activates PKC and IP3, which induce sustained calcium mobilization. It is striking that patients with mutations in Btk do not have clinical abnormalities in platelet or myeloid cell function. It is possible that activation of Btk in platelets, which results in increased platelet aggregation and dense granule secretion, or in mast cells, which enhances histamine release and cytokine production, results in an inflammatory reaction that is no longer clinically advantageous.
Patients with XLA are usually recognized to have immunodeficiency when they are hospitalized for a dramatic infection such as sepsis, meningitis or pneumonia with empyema.27 Because XLA is maintained in the population by new mutations in Btk, only about half of the affected patients have a family history of disease.5,10 The mean age at diagnosis in patients with sporadic XLA is 30 to 40 months. Patients with affected family members are generally recognized to have antibody deficiency at an earlier age, but it is only in the last 10 years that patients with affected brothers, uncles or cousins have been evaluated for immunodeficiency before they develop symptoms. Patients who are diagnosed as having XLA at greater than one year of age almost always have a history of chronic otitis.27 Palpation of cervical lymph nodes and an awareness that the absence of lymph nodes is pathologic could enhance early recognition of patients with chronic otitis and XLA.
XLA is a profound but leaky defect in B cell development. Most patients demonstrate markedly reduced concentrations of serum IgG, IgM, IgA and IgE. However, almost all patients do have some measurable IgG and occasionally patients will have some IgM and IgA.28,29 Antibodies to vaccines and infectious organisms are absent. The most consistent and distinctive finding in patients with XLA is the profound reduction in the number of B cells in the peripheral circulation.28,30 In the normal individual between 5% and 15% of the peripheral blood lymphocytes are CD19+ B cells. The mean number of CD19+ B cells in patients with XLA who are less than 10 years of age is 0.1%; older patients with XLA tend to have fewer B cells. Analysis of B cell precursors in the bone marrow demonstrates normal numbers of pro-B cells, cells that are positive for CD34 and CD19, but very few pre-B cells or mature B cells.30,31 The transition from pro-B cell to pre-B cells requires signaling through the pre-B cell receptor. Thus, the block at this stage of differentiation in patients with XLA provides strong evidence that Btk is involved in signaling through the pre-B cell receptor.
About 20% of patients with sporadic XLA are recognized to have immunodeficiency at less than a year of age when they are hospitalized with cellulitis, abscess or pyoderma gangrenosum; staphylococcal or pseudomonas sepsis; and neutropenia.27 This constellation of findings is less common as a presenting finding in older patients with XLA but it does occur. The presence of Btk in myeloid cells could suggest that Btk has a role in control of neutrophil production or response to stress; however, the fact that other patients with profound defects in B cell production, for example, patients with μ heavy chain defect, have a similar incidence of neutropenia and pseudomonas or staphylococcal sepsis makes this unlikely. The neutropenia in patients with severe defects in B cell development resolves within 3 to 6 weeks of the initiation of gammaglobulin replacement therapy and it is not seen in patients receiving adequate therapy. A plausible explanation for the neutropenia is that the patients acquire a viral infection and develop secondary neutropenia; the neutropenia then leaves the patients vulnerable to staphylococcal and pseudomonas infections, infections that are typical of neutropenia.
The majority of patients with XLA are recognized to have immunodeficiency at 1 to 4 years of age, but about 10% of patients are more than 5 years old at the time of diagnosis. Some but not all of the patients who are diagnosed at an older age have higher concentrations of serum immunoglobulins. Acute life-threatening infections are uncommon in XLA patients receiving gammaglobulin but many patients continue to have problems with otitis or sinusitis, and 10% to 20% of patients develop significant problems despite optimal therapy.32 Patients who have pulmonary scarring at the time of diagnosis are more likely to have continuing episodes of pneumonia and bronchitis.29,33 In addition to H. influenzae and S. pneumoniae, patients with XLA are more susceptible to infection with mycoplasmas and ureoplasmas. These organisms, which are difficult to culture and difficult to eradicate, may cause chronic, persistent pneumonia, arthritis, cystitis or cellulitis.34,35 Like all patients with hypogammaglobulinemia, patients with XLA have an increased incidence of giardiasis.36 Inflammatory bowel disease has been reported in these patients37 and, surprisingly, some patients with XLA have what appear to be seasonal allergies or drug allergies.
It has been recognized for over 30 years that patients with profound hypogammaglobulinemia and absent B cells are particularly susceptible to chronic enteroviral infections, including vaccine associated polio.38,39 ECHO 11 appears to be the most common organism, although other ECHO strains and coxsackievirus have been reported.40 The enteroviral infections are often insidious in onset and may present with ataxia, behavioral changes, headaches or dermatomyositis-like symptoms. The course of this devastating complication tends to wax and wane but is often ultimately fatal. High-dose intravenous gammaglobulin and intrathecal gammaglobulin have been used, but they do not necessarily result in long-term improvements. Prognosis may be influenced by the duration of infection before high-dose gammaglobulin is started. It should be noted that it can be very difficult to document the enteroviral infection; however, polymerase chain reaction (PCR) analysis of cerebral spinal fluid may be helpful.41 The incidence of chronic enteroviral infection has decreased but not disappeared since the mid-1980s when most patients were switched from plasma therapy or intramuscular gammaglobulin to intravenous gammaglobulin. It is possible that patients who have developed signs of enteroviral infection while receiving intravenous gammaglobulin actually acquired the infection before the onset of treatment.
The long-term prognosis in patients with XLA continues to improve. Before 1960 the majority of patients died of acute or chronic infections in early childhood. In the 1970s, a significant proportion of patients died of chronic enteroviral infections or progressive pulmonary disease before they reached adulthood.5 However, in the last 10 to 20 years, the majority of patients with XLA have reached adulthood with few significant problems. As is true with all patients who have the onset of a chronic disease in childhood, care must be taken to avoid or treat psycho-social problems and encourage the development of independence.
Almost every family with XLA has a “private mutation” in Btk. Over 400 different mutations have been identified and no single mutation accounts for more than 5% of the patients.10,42 The majority of mutations are single base pair substitutions resulting in premature stop codons, splice defects or amino acid substitutions. About 20% of mutations consist of an insertion or deletion of 1–5 base pairs; these mutations generally cause a frameshift and a secondary premature stop codon. Gross alterations of the gene, including large deletions, duplications, insertions or inversions account for the remaining 5% of mutations.10 Most often, mutation detection is performed using a screening evaluation that analyzes genomic DNA exon by exon, followed by DNA sequencing of a region found to be abnormal; however, no single technique allows the identification of all mutations. For example, Southern blot analysis or sequencing would be required to detect a duplication.43
Premature stop codons, frameshift mutations and splice defects in Btk generally result in faulty processing of the Btk message and an absence of transcripts in the cytoplasm.19 In addition, a high proportion of amino acid substitutions in Btk render the protein unstable.44– 46 Thus, the majority of mutations in Btk result in markedly reduced or absent Btk protein. Although some mutations in Btk may be more likely to give rise to higher concentrations of serum immunoglobulins or delayed detection of disease, the genotype/phenotype correlation in XLA is weak.
Autosomal Recessive Agammaglobulinemia
Patients with autosomal recessive agammaglobulinemia due to mutations in μ heavy chain, λ5, Igα or BLNK are not easily distinguished from patients with XLA by routine clinical or laboratory studies. Fourteen families with defects in μ heavy chain have been reported;9,11,12 in all cases, if the first affected family member was a male, he was diagnosed as having XLA. An alternative diagnosis was considered only when a female family member was identified or molecular studies did not demonstrate a mutation in Btk. Nevertheless, when the group of patients with defects in μ heavy chain are compared to the patients with XLA, the onset of disease is earlier and the incidence of severe complications is higher in the patients with defects in μ heavy chain.9 The mean age at diagnosis in patients with mutations in μ heavy chain was 11 months, whereas the mean age at diagnosis in patients with XLA was 35 months. Seven of the 19 patients with documented mutations in μ heavy chain had significant enteroviral infections and 6 of the 19 had severe neutropenia. These differences may be due to the fact that μ heavy chain defect causes a complete block at the pro-B cell to pre-B cell transition rather than a leaky block as is seen in XLA.
Ten different mutations in μ heavy chain have been reported; half of these mutations are large deletions that remove not only the μ constant region gene but also most of the D and J region genes.9 Of the remaining 5 mutations, 2 were frameshift mutations, 1 was a premature stop codon, 1 was an amino acid substitution at an invariant cysteine and the last was a base pair substitution at the splice site required to make the membrane form of μ heavy chain. This last mutation was seen in 6 different families from 4 different countries. Analysis of polymorphic markers within the immunoglobulin locus indicated that the splice site mutation occurred on at least 3 different haplotypes, marking this site as a hot spot for mutations in μ heavy chain.
One patient with compound heterozygous mutations in λ5,13 3 patients with defects in Igα,14,15 and 2 patients with mutations in BLNK have been identified.16 These patients also had clinical and laboratory findings consistent with the diagnosis of XLA. Over 75% of the patients with autosomal recessive agammaglobulinemia were homozygous for their gene defect and most were from families that had known consanguinity or were from relatively isolated populations.
Hyper IgM Syndrome
The term hyper IgM syndrome is used to describe a heterogeneous group of disorders characterized by recurrent bacterial infections and very low or absent serum IgG, IgA and IgE.7 Serum IgM may be normal or elevated. The number of B cells in the peripheral circulation is generally normal. About 65% of children with hyper IgM syndrome are males with mutations in CD40 ligand (CD154),3,47 a gene encoded on the long arm of the X chromosome. CD40 ligand is a member of the tumor necrosis factor (TNF) family that is transiently expressed on the surface of activated CD4+ T cells. Its cognate receptor, CD40, is expressed on B cells, macrophages, dendritic cells and some endothelial, epithelial and carcinoma cells. Stimulation of B cells through CD40 can activate proliferation or apoptosis, depending on the stage of differentiation of the B cell. Ligation of CD40 is also essential for isotype switch to IgG, IgA and IgE.
Patients with defects in CD40 ligand have recurrent bacterial infections, like other patients with antibody deficiency, but they also have opportunistic infections.8 As many as 30% of patients are recognized to have immunodeficiency when they present with pneumocystis pneumonia in the first year of life. Cryptosporidium, toxoplasmosis and atypical mycobacterial infections can also be troublesome. The expanded spectrum of infections in patients with X-linked hyper IgM syndrome can be explained by the fact that activated T cells expressing CD40 ligand stimulate CD40-bearing dendritic cells and monocytes. These cells then enhance cytotoxic T cell function and secretion of cytokines, particularly interleukin (IL)-12.48,49 IL-12, in turn, stimulates T cells to produce gamma interferon.50 The absence of this crosstalk results in a defective T cell response to pathogens and impaired T cell maturation.
Neutropenia, often associated with superficial mouth ulcers, is seen in over 50% of patients with mutations in CD40 ligand.8 In some patients the neutropenia is chronic and severe, in others it is intermittent. B cells, thymic epithelial cells, and perhaps other cells secrete granulocyte colony-stimulating factor (G-CSF) in response to CD40 stimulation.51,52 This helps explain the neutropenia in patients with CD40 ligand deficiency, but it is not clear why some but not all patients have this finding. The occurrence of neutropenia is not associated with the specific mutation in CD40 ligand. Patients with defects in CD40 ligand also have an elevated incidence of carcinomas of the liver, pancreas and biliary tree.53 Ligation of CD40 on activated, infected or inflamed epithelial cells helps protect these cells from uncontrolled proliferation.
Four patients with mutations in the gene for CD40 have been reported.54,55 These patients are clinically indistinguishable from the patients with mutations in CD40 ligand, suggesting that this ligand and its receptor do not interact with other partners. Patients with CD40 ligand deficiency have been successfully treated with bone marrow transplantation.56– 58 Because CD40 is expressed on non-hematopoietic cells, it is less likely that patients with defects in the gene for this protein will benefit from hematopoietic stem cell transplants.
The most common form of autosomal recessive hyper IgM syndrome is due to mutations in a gene encoded at 12p13, activation induced cytidine deaminase (AID).59,60 AID is required for normal isotype switch and hyper mutation of immunoglobulin variable region genes.61 Activation of B cells by CD40 ligation and/or other stimuli induces the expression of AID, which functions by deaminating cytosines within the immunoglobulin locus.62 The resulting uracil is removed, causing a break in the DNA, or repaired in an error prone fashion. Patients with defects in AID are not as sick as patients with early defects in B cell differentiation. Almost half of the patients are recognized to have immunodeficiency at greater than 10 years of age. These patients have a markedly elevated incidence of lymphoid hyperplasia.
A second form of X-linked hyper IgM syndrome is caused by mutations in a component of the NF-κB pathway, NEMO.63–,65 NEMO, also known as IKKγ, acts as a scaffold to bind 2 proteins with kinase activity: IKKα and IKKβ. Together IKKα and IKKβ and IKKγ phosphorylate I-κB, a molecule that sequesters NF-κB in the cytoplasm. When I-κB is phosphorylated, it releases NF-κB, allowing it to move into the nucleus and act as a transcription factor. Most mutations in NEMO are embryonically lethal for the male and cause incontinentia pigmenti in heterozygous females;66 however, milder mutations in NEMO can result in a variable phenotype, including hyper IgM syndrome, in affected males. Most affected males have abnormal, conical teeth, inadequate sweating and poor antibody production to polysaccharide antigens. Not all males with defects in NEMO have low IgG and IgA.
Conclusions
Single gene defects of the immune system resulting in antibody deficiencies are rare but fascinating disorders that provide valuable insights into development of the normal immune system. In patients with early defects in B cell development or mutations in CD40 ligand or CD40, neutropenia may be the presenting finding. Thus, the threshold for measuring serum immunoglobulins should be low in any child with neutropenia. Patients with antibody deficiencies should be treated with gammaglobulin replacement and referral to a center with expertise in immunodeficiency.
III. Severe Combined Immunodeficiency Update
Jennifer M. Puck, MD*
Chief, Genetics and Molecular Biology Branch, NHGRI/NIH, 49 Convent Drive, Building 49, Room 4A14, Bethesda MD 20892
Severe combined immunodeficiency (SCID) is a term used to describe a collection of genetic defects in both humoral and cellular immunity that have an early clinical presentation and, if untreated, a fatal outcome in the first few years of life. The incidence of SCID is unknown, but it is estimated to be in 1 per 50,000 to 100,000 births in all ethnic groups. One reason for the lack of accurate incidence data is that infants with SCID may die of infections without having the condition recognized. The profound degree of immune compromise in SCID leads to infections with bacterial, viral, and fungal pathogens that cause failure to thrive and chronic diarrhea. The infections are not only frequent, but may also be severe; persistent despite standard medical treatment; recurrent; and caused by opportunistic pathogens. Immunological and infectious characteristics of SCID are summarized in Table 4 .1,2
Congenital immunodeficiency causing early fatality was first reported in the 1950s in Switzerland. Hitzig and Willi,3 following the discovery of agammaglobulinemia by Bruton,4 reported familial alymphocytosis and agammaglobulinemia with a fatal outcome in infancy. The syndrome was called Swiss-type agammaglobulinemia to distinguish the very severely affected Swiss patients, who in retrospect lacked both B-cell and T-cell function, from patients such as the one Bruton reported who lacked only B cells. The confusing term “Swiss-type agammaglobulinemia” was applied to patients with X-linked SCID (XSCID) for many years, but has been replaced with XSCID, SCIDX1, or gamma-c (γc) deficient SCID after the X-linked gene defect became known.5,6 Adenosine deaminase (ADA) deficiency was serendipitously associated with SCID by Giblett et al in 19727 (reviewed in Hirschhorn8). Many additional gene defects are known to produce SCID, and new SCID genotypes are identified each year.9,10 Today, there are improved immunological tools to characterize immune defects as well as the ability to identify the genes that are mutated in specific forms of SCID. Current disease designations for SCID are summarized in Table 5 . XSCID is the most frequent type, accounting for around half of all SCID cases and explaining the preponderance of males with SCID. This is followed in frequency by ADA deficiency and then interleukin (IL)-7 receptor alpha (IL7RA) chain deficiency and Janus kinase 3 (JAK3) deficiency, after which a host of more rare genetic defects account for smaller fractions of the total incidence.10– 15
Despite great advances in the understanding of genetic causes of SCID, failure of physicians to consider immunodeficiency is a major hurdle to rescuing affected infants. Its frequency is so low that most family doctors will never observe a case. Even today, SCID is frequently not suspected early, even when clinical clues are available. Family history of infant deaths in past generations and appreciation of low total lymphocyte counts could help increase early diagnosis. Normal newborn mean values for total lymphocyte number, 5400/μL, and T-cell number, 3100/μL,16 are about twice the normal adult values most medical professionals are more familiar with.
Prior to 1968, patients with SCID almost always died within the first year of life. However, the achievement of immune reconstitution following bone marrow transplantation from an HLA-identical sibling marked the beginning of successful treatment.17 The unavailability of a histocompatible donor led to the use of isolation in a gnotobiotic environment for David, the “Bubble Boy,” the famous Texas patient who came to represent SCID to the general public. Although David succumbed to Epstein-Barr virus lymphoproliferation after a haploidentical T-cell depleted transplant at age 12 from a sibling,18 the technique of T-cell depletion has now been successfully adopted for treatment of infants with SCID. Early diagnosis, better antibiotics, and intensive supportive care have also improved treatment of SCID over the past 3 decades. Currently it is estimated that 75% to greater than 90% of infants diagnosed with SCID are saved by hematopoietic stem cell transplantation treatment, with better survival for transplants from HLA-matched related donors.15,19,20
XSCID will be discussed in some detail as the most common type of SCID and the one for which there is the most data about treatments, including gene therapy. Only males, who have a single X chromosome, are affected with XSCID. Female carriers, with a mutated copy of the gene on 1 and a normal copy on the other of their 2 X chromosomes, are healthy but pass on the condition to 50% of their male offspring. Patients with XSCID generally have very few T cells, but normal or increased numbers of functionally impaired B cells (T−B+ SCID). They lack natural killer (NK) cells. Occasional XSCID patients have substantial numbers of maternally derived T cells or allogeneic lymphocytes acquired through non-irradiated transfusions, so that T cell lymphopenia is not always present. In 1993, Noguchi et al5 and Puck et al6 discovered that XSCID is caused by mutations in IL2RG, discovered in 1992 to be the gene encoding the third chain or γ chain of the IL-2 receptor.21 Subsequent appreciation that the γ protein was part of several transmembrane receptors for cytokines (including IL-4, IL-7, IL-9, IL-15, IL-21) led to naming the protein the common γ chain (γc). The protein is a member of the cytokine receptor gene superfamily and is encoded by a gene of 8 exons. When engaged by extracellular cytokine ligation, the intracellular portion of γc interacts with JAK3, resulting in phosphorylation of intracellular signaling kinases and signal translators and activators of transcription (STATs), to modify the cell’s transcription program. Like every primary immunodeficiency to date, XSCID does not have a predominant mutation, but rather hundreds of mutations, many unique, throughout its coding and regulatory regions. Most are small changes, deletions, or insertions on the order of one to a few nucleotides.2,22 Many mutations decrease RNA stability, and almost all result in complete abolition of signaling. However, some mutations, such as truncations of the intracellular domain, give rise to proteins that are expressed on the cell surface, but are nonfunctional. Thus immunofluorescence staining for absence of γc is not a reliable diagnostic method for XSCID.
The best current treatment for XSCID is transplantation of hematopoietic stem cells (either from bone marrow or from peripheral blood after mobilization) from an HLA-matched relative. Unfortunately, most patients lack a matched related donor, but haploidentical, T-cell depleted transplants and transplants from unrelated matched donors have been used with success. Survival rates in this previously fatal disease are above 90% in some series.15 Chemotherapy prior to transplant is used by some centers, but because the host is immunodeficient, this is not necessary for rescuing T-cell function. Although some patients have fully functional immunity following their transplant, others fail to have B-cell function restored and some lose their grafts over time, suggesting that true stem cells may not have been established from the donor. These considerations have led to the pursuit of gene transfer into autologous cells, by means of retroviruses, to treat XSCID.
For several reasons, as outlined in Table 6 , XSCID appeared to be a good candidate disease for retroviral gene therapy. Ubiquitous expression of IL2RG mRNA has been demonstrated in mouse and human hematopoietic lineages.24,25 The fact that there are relatively constant, substantial levels of mRNA even in the most immature bone marrow fractions suggests that expression using strong promotors in retroviral constructs may not be harmful. Importantly, there is a natural selective advantage for lymphocytes expressing wild type γc, as demonstrated by the skewed X-chromosome inactivation in lymphocyte lineages of female carriers.26 Moreover, natural reversion to normal of a SCID-causing IL2RG mutation has been reported in a T lymphoid precursor that expanded to provide a diverse repertoire and modify the SCID phenotype.27 In preclinical studies, retroviral transduction of IL2RG cDNA constructs brought about expression of normal γc in B-cell lines from XSCID patients and corrected their defects in IL-2– and IL-4–mediated phosphorylation of JAK3.28,29 XSCID patient stem cells transduced with the normal gene became able to differentiate into T and B cells in a chimeric sheep model.30 The major limitation of retroviral gene therapy has been the poor efficiency of transduction into self-renewing bone marrow hematopoietic stem cell populations. However, cytokine activation in vitro can increase transduction rates by retroviruses. Given encouraging preliminary data, human gene therapy trials were undertaken and have achieved successful XSCID immune reconstitution.31,32 Nine of 10 infants followed up in this trial developed new T cells bearing the correct copy of the IL2RG cDNA as detected by PCR. Corrected B and NK cells were also detectable, and antibody responses to childhood vaccines were in some of the children completely normal. Children were able to live free of infections without continuous antibiotics or immunoglobulin supplementation, making XSCID the first human disease to be successfully treated solely by gene therapy. Unfortunately, 2 of the patients in this original series developed leukemic expansions of T-cell clones 27 and 30 months following their gene therapy. These were the 2 youngest infants treated (Table 6 ). Insertion of the retroviral vector in 2 different locations near the 5′ end of the LIM domain only-2 (LMO-2) transcription factor with inappropriate expression of LMO-2 characterized the proliferating cells in both children. This unanticipated complication has led to intensive efforts to understand the spectrum of retroviral insertions in humans, the relationship of LMO-2 expression to pathways involving γc, and whether XSCID is a uniquely high-risk disease for gene therapy or if the risk of leukemia is present in all SCID genotypes and perhaps other conditions approached by retroviral gene therapy. It is interesting to note that a small series of infants treated for ADA SCID with gene therapy preceded by nonablative conditioning appears to have also achieved good immune reconstitution.33 More time and larger numbers of patients will be needed to determine the risks and benefits of gene therapy, which is presently in its infancy.
The dramatic turnaround in outlook for patients with SCID and their families over the past generation is one of the most striking in modern medicine. A previously uniformly fatal disease is now effectively treated by stem cell transplantation, the cause and pathogenesis of the disease can be known in the majority of cases, specific gene defects can be identified and traced in patients and at-risk family members, and gene therapy is a potential option for treatment.
Immune receptors encoded by genes in the leukocyte receptor complex region on human chromosome 19q13.4. Common names and CD designations of the receptors are indicated. Functions and associations with adapter proteins are implied based on structural characteristics of the receptors, but have only been validated experimentally for a few of the receptors, usually when a ligand has been identified. As indicated, certain KIR2DL recognize a dimorphic polymorphism in HLA-C at positions 77 and 80 in the α1 helix of the class I heavy chain. N77K80 and S77N80 refer to the amino acids (single letter code) present at residues 77 and 80 in HLA-C, conferring specificity for the indicated KIR. * = KIR2DL2 and KIR2DL3 are alleles of the same locus; KIR3DL1 and KIR3DS1 are alleles of the same locus. All other receptors listed are encoded by separate genes, although many of these genes also demonstrate allelic polymorphism.
| Names . | CD . | Function . | Signaling . | Ligand . |
|---|---|---|---|---|
| ILT5 / LIR3 / LILRB3 | CD85a | Inhibition | ITIM | |
| ILT8 / LILRB6 | CD85b | Activation | FcεRIγ | |
| LIR8 / LILRB5 | CD85c | Inhibition | ITIM | |
| ILT4 / LIR2 / LILRB2 | CD85d | Inhibition | ITIM | HLA class I |
| ILT6 / LIR4 / LILRA3 | CD85e | Soluble | - | |
| ILT11 / LILRB7 | CD85f | Activation | FcεRIγ | |
| ILT7 / LILRA4 | CD85g | Activation | FcεRIγ | |
| LAIR-1 | Inhibition | ITIM | ||
| LAIR-2 | Soluble receptor | |||
| ILT1 / LIR7 / LILRA2 | CD85h | Activation | FcεRIγ | |
| LIR6 / LILRA1 | CD85i | Activation | FcεRIγ | |
| ILT2 / LIR1/ LILRB1 | CD85j | Inhibition | ITIM | HLA class I, CMV UL18 |
| ILT3 / LIR5/ LILRB4 | CD85k | Inhibition | ITIM | |
| ILT9 / LILRA6 | CD85l | Pseudogene | ||
| ILT10 / LILRA5 | CD85m | Activation | FcεRIγ | |
| KIR3DL3 | CD158z | Inhibition | ITIM | |
| KIR2DL3* | CD158b2 | Inhibition | ITIM | HLA-C S77/N80 |
| KIR2DL2* | CD158b1 | Inhibition | ITIM | HLA-C S77/N80 |
| KIR2DL1 | CD158a | Inhibition | ITIM | HLA-C N77/K80 |
| KIR2DL4 | CD158d | Inhibition/ Activation? | ITIM / ? | |
| KIR3DL1* | CD158e1 | Inhibition | ITIM | HLA-Bw4 |
| KIR3DS1* | CD158e2 | Activation | DAP12 | |
| KIR2DL5A | CD158f | Inhibition | ITIM | |
| KIR2DL5B | Inhibition | ITIM | ||
| KIR2DS3 | Activation | DAP12 | ||
| KIR2DS5 | CD158g | Activation | DAP12 | |
| KIR2DS1 | CD158h | Activation | DAP12 | HLA-C, weakly |
| KIR2DS2 | CD158j | Activation | DAP12 | |
| KIR2DS4 | CD158i | Activation | DAP12 | HLA-C, weakly |
| KIR3DL2 | CD158k | Inhibition | ITIM | HLA-A? |
| FcαR | CD89 | Activation | FcεRIγ | IgA |
| NKp46 | Activation | FcεRIγ, ζ |
| Names . | CD . | Function . | Signaling . | Ligand . |
|---|---|---|---|---|
| ILT5 / LIR3 / LILRB3 | CD85a | Inhibition | ITIM | |
| ILT8 / LILRB6 | CD85b | Activation | FcεRIγ | |
| LIR8 / LILRB5 | CD85c | Inhibition | ITIM | |
| ILT4 / LIR2 / LILRB2 | CD85d | Inhibition | ITIM | HLA class I |
| ILT6 / LIR4 / LILRA3 | CD85e | Soluble | - | |
| ILT11 / LILRB7 | CD85f | Activation | FcεRIγ | |
| ILT7 / LILRA4 | CD85g | Activation | FcεRIγ | |
| LAIR-1 | Inhibition | ITIM | ||
| LAIR-2 | Soluble receptor | |||
| ILT1 / LIR7 / LILRA2 | CD85h | Activation | FcεRIγ | |
| LIR6 / LILRA1 | CD85i | Activation | FcεRIγ | |
| ILT2 / LIR1/ LILRB1 | CD85j | Inhibition | ITIM | HLA class I, CMV UL18 |
| ILT3 / LIR5/ LILRB4 | CD85k | Inhibition | ITIM | |
| ILT9 / LILRA6 | CD85l | Pseudogene | ||
| ILT10 / LILRA5 | CD85m | Activation | FcεRIγ | |
| KIR3DL3 | CD158z | Inhibition | ITIM | |
| KIR2DL3* | CD158b2 | Inhibition | ITIM | HLA-C S77/N80 |
| KIR2DL2* | CD158b1 | Inhibition | ITIM | HLA-C S77/N80 |
| KIR2DL1 | CD158a | Inhibition | ITIM | HLA-C N77/K80 |
| KIR2DL4 | CD158d | Inhibition/ Activation? | ITIM / ? | |
| KIR3DL1* | CD158e1 | Inhibition | ITIM | HLA-Bw4 |
| KIR3DS1* | CD158e2 | Activation | DAP12 | |
| KIR2DL5A | CD158f | Inhibition | ITIM | |
| KIR2DL5B | Inhibition | ITIM | ||
| KIR2DS3 | Activation | DAP12 | ||
| KIR2DS5 | CD158g | Activation | DAP12 | |
| KIR2DS1 | CD158h | Activation | DAP12 | HLA-C, weakly |
| KIR2DS2 | CD158j | Activation | DAP12 | |
| KIR2DS4 | CD158i | Activation | DAP12 | HLA-C, weakly |
| KIR3DL2 | CD158k | Inhibition | ITIM | HLA-A? |
| FcαR | CD89 | Activation | FcεRIγ | IgA |
| NKp46 | Activation | FcεRIγ, ζ |
Human Fc receptors: Common names, functions and distribution.
| Fc Receptor . | Ig Specificity . | Function . | Signaling . | Distribution . |
|---|---|---|---|---|
| CD16a, FcγRIII | IgG | Activation | FcεRIγ, ζ | NK, macrophage, T subset |
| CD16b, FcγRIII | IgG | ? | Neutrophils | |
| CD32a, FcγRIIA | IgG | Activation | Intrinsic ITAM | Myeloid, B |
| CD32b1, FcγRIIB | IgG | Inhibitory | ITIM | Myeloid, B |
| CD64, FcγRI | IgG | Activation | FcεRIγ | Myeloid |
| CD89, FcαRI | IgA | Activation | FcεRIγ | Myeloid |
| Polymeric Ig | IgM, IgA | Transcytosis | Epithelial cells | |
| Fcα/μR | IgM, IgA | Endocytosis | Macrophage, B | |
| FcεRI | IgE (high affinity) | Activation | FcεRIγ, FcεRIβ | Mast cells, basophils |
| FcRn | IgG | Transcytosis | Epithelial cells, myeloid | |
| CD23 | IgE (low affinity | Endocytosis | B |
| Fc Receptor . | Ig Specificity . | Function . | Signaling . | Distribution . |
|---|---|---|---|---|
| CD16a, FcγRIII | IgG | Activation | FcεRIγ, ζ | NK, macrophage, T subset |
| CD16b, FcγRIII | IgG | ? | Neutrophils | |
| CD32a, FcγRIIA | IgG | Activation | Intrinsic ITAM | Myeloid, B |
| CD32b1, FcγRIIB | IgG | Inhibitory | ITIM | Myeloid, B |
| CD64, FcγRI | IgG | Activation | FcεRIγ | Myeloid |
| CD89, FcαRI | IgA | Activation | FcεRIγ | Myeloid |
| Polymeric Ig | IgM, IgA | Transcytosis | Epithelial cells | |
| Fcα/μR | IgM, IgA | Endocytosis | Macrophage, B | |
| FcεRI | IgE (high affinity) | Activation | FcεRIγ, FcεRIβ | Mast cells, basophils |
| FcRn | IgG | Transcytosis | Epithelial cells, myeloid | |
| CD23 | IgE (low affinity | Endocytosis | B |
Single gene defects of the immune system resulting in antibody deficiencies.
| Agammaglobulinemia with absent B cells | |
| X-linked agammaglobulinemia (Bruton’s) | 85% |
| μ heavy chain | 5% |
| Surrogate light chain | < 1% |
| Igα/Igβ | 2% |
| BLNK | 2% |
| Hyper IgM syndrome | |
| CD40 ligand | 65% |
| CD40 | < 5% |
| AID | 15% |
| NEMO | 10% |
| Agammaglobulinemia with absent B cells | |
| X-linked agammaglobulinemia (Bruton’s) | 85% |
| μ heavy chain | 5% |
| Surrogate light chain | < 1% |
| Igα/Igβ | 2% |
| BLNK | 2% |
| Hyper IgM syndrome | |
| CD40 ligand | 65% |
| CD40 | < 5% |
| AID | 15% |
| NEMO | 10% |
Infectious and immunological characteristics
| Clinical clues |
| Persistent infections despite standard treatment |
| Recurrent infections |
| Severe infections, including invasive infections such as meningitis, septic arthritis |
| Infections with opportunistic pathogens |
| Graft vs host disease either from maternally acquired lymphocytes or nonirradiated transfusions |
| Absent thymic shadow on chest x-ray |
| Family history of immunodeficiency or infant deaths from infection |
| Infectious agents |
| Bacteria, including Gram positive, Gram negative |
| Fungi |
| Viruses |
| Rotavirus and other intestinal viruses |
| Respiratory syncytial virus and other respiratory viruses |
| Cytomegalovirus and other herpesviruses |
| Opportunistic agents, such as Pneumocystis and disseminated bacillus Calmette-Guerin (BCG) (in countries using neonatal BCG vaccination) |
| Immunologic abnormalities |
| T cell lymphopenia < 2200 (compared with age-matched control values—healthy infants have higher lymphocyte counts than older individuals) |
| Poor responses to T-cell mitogens and antigens |
| Absent or nonfunctional B cells |
| Falling immunoglobulin (Ig) G (with waning maternal transplacental antibodies) and very low levels of IgA, IgM |
| Absent antibody responses to specific antigens, such as tetanus toxoid |
| Clinical clues |
| Persistent infections despite standard treatment |
| Recurrent infections |
| Severe infections, including invasive infections such as meningitis, septic arthritis |
| Infections with opportunistic pathogens |
| Graft vs host disease either from maternally acquired lymphocytes or nonirradiated transfusions |
| Absent thymic shadow on chest x-ray |
| Family history of immunodeficiency or infant deaths from infection |
| Infectious agents |
| Bacteria, including Gram positive, Gram negative |
| Fungi |
| Viruses |
| Rotavirus and other intestinal viruses |
| Respiratory syncytial virus and other respiratory viruses |
| Cytomegalovirus and other herpesviruses |
| Opportunistic agents, such as Pneumocystis and disseminated bacillus Calmette-Guerin (BCG) (in countries using neonatal BCG vaccination) |
| Immunologic abnormalities |
| T cell lymphopenia < 2200 (compared with age-matched control values—healthy infants have higher lymphocyte counts than older individuals) |
| Poor responses to T-cell mitogens and antigens |
| Absent or nonfunctional B cells |
| Falling immunoglobulin (Ig) G (with waning maternal transplacental antibodies) and very low levels of IgA, IgM |
| Absent antibody responses to specific antigens, such as tetanus toxoid |
| Defects in Cytokine Receptors and Cytokine Signaling (B cells generally present but nonfunctional) |
| X-linked SCID, due to defects in the γc chain found in receptors for interleukin (IL)-2, -4, -7, -9, -15, and -21 |
| JAK3 deficiency, due to defects in the Janus signaling kinase that interacts with the intracellular portion of the common gamma chain |
| IL-7 receptor alpha chain deficiency, due to defects in the receptor for IL-7, critical for lymphoid cell development and differentiation |
| CD45 deficiency, due to defects in a transmembrane tyrosine phosphatase |
| Defects in Recombination of the Antigen Receptor Genes of B cells and T cells (T and B cells absent) |
| Recombinase activating gene 1 (RAG1) deficiency |
| Recombinase activating gene 2 (RAG2) deficiency |
| Artemis deficiency (another component of the recombinase system) |
| Defects in purine pathway enzymes that result in buildup of metabolites toxic to lymphocytes |
| Adenosine deaminase (ADA) deficiency |
| Purine nucleoside phosphorylase (PNP) deficiency |
| Other lymphocyte specific gene disorders (may have partial preservation of some lymphocyte lineages/function) |
| CD3 deficiency |
| ZAP70 deficiency |
| MHC Class II deficiency due to diverse gene defects in transcription factors for the Class II molecules |
| MHC Class I deficiency due to defects in TAP1/TAP2, transporters involved in Class I expression |
| SCID phenotype as a component of syndromes (severity of disease may be variable) |
| Cartilage hair hypoplasia, due to defects in RMRP, an RNA component of a ribonucleoprotein RNAse |
| Severe DiGeorge syndrome, in 90% of cases associated with heterozygous deletion of chromosome 22q11 |
| SCID with alopecia and nail dystrophy (orthologous to nude mouse) due to defects in winged helix nude (WHN) transcription factor |
| SCID with multiple bowel atresias |
| Reticular dysgenesis, a stem cell defect with impaired maturation of multiple bone marrow lineages |
| Defects in Cytokine Receptors and Cytokine Signaling (B cells generally present but nonfunctional) |
| X-linked SCID, due to defects in the γc chain found in receptors for interleukin (IL)-2, -4, -7, -9, -15, and -21 |
| JAK3 deficiency, due to defects in the Janus signaling kinase that interacts with the intracellular portion of the common gamma chain |
| IL-7 receptor alpha chain deficiency, due to defects in the receptor for IL-7, critical for lymphoid cell development and differentiation |
| CD45 deficiency, due to defects in a transmembrane tyrosine phosphatase |
| Defects in Recombination of the Antigen Receptor Genes of B cells and T cells (T and B cells absent) |
| Recombinase activating gene 1 (RAG1) deficiency |
| Recombinase activating gene 2 (RAG2) deficiency |
| Artemis deficiency (another component of the recombinase system) |
| Defects in purine pathway enzymes that result in buildup of metabolites toxic to lymphocytes |
| Adenosine deaminase (ADA) deficiency |
| Purine nucleoside phosphorylase (PNP) deficiency |
| Other lymphocyte specific gene disorders (may have partial preservation of some lymphocyte lineages/function) |
| CD3 deficiency |
| ZAP70 deficiency |
| MHC Class II deficiency due to diverse gene defects in transcription factors for the Class II molecules |
| MHC Class I deficiency due to defects in TAP1/TAP2, transporters involved in Class I expression |
| SCID phenotype as a component of syndromes (severity of disease may be variable) |
| Cartilage hair hypoplasia, due to defects in RMRP, an RNA component of a ribonucleoprotein RNAse |
| Severe DiGeorge syndrome, in 90% of cases associated with heterozygous deletion of chromosome 22q11 |
| SCID with alopecia and nail dystrophy (orthologous to nude mouse) due to defects in winged helix nude (WHN) transcription factor |
| SCID with multiple bowel atresias |
| Reticular dysgenesis, a stem cell defect with impaired maturation of multiple bone marrow lineages |
X-linked severe combined immunodeficiency as a candidate disease for gene therapy.
| *Data from Kohn et al23; A. Fischer and M. Cavazzana-Calvo, personal communication in oral presentations, 2003 |
| Advantages |
| Complications and late failures after allogeneic transplantation mandate a search for better treatment. |
| Ex-vivo transduction: hematopoietic cells can be removed and enriched, cultured with retrovirus and then reinfused intravenously, where they will home to the bone marrow. |
| Disease gene product, the common gamma chain of cytokine receptors is widely expressed in blood lineages; overexpression apparently not harmful. |
| The problem of immune elimination of the corrected cells should not arise because of the immunodeficiency phenotype. |
| There is an in vivo selective advantage for lymphoid cells expressing the normal common gamma chain protein. |
| Animal models and human trials have been successful in restoring immunocompetence. |
| Of 10 infants given gene therapy as primary treatment in France by the group of Fischer et al, 9 had restoration of T-cell function.31,32 |
| Specific antibody production was restored in most. |
| A dose of > 1 million corrected CD34+ cells/kg was established as required to achieve full reconstitution. |
| Disadvantages* |
| Serious adverse events occurred in 2/10 infants: development of clonal leukemic proliferations of transduced cells. |
| Both were the youngest infants at treatment (1 and 3 months). |
| Clonal proliferations were detected about 2½ years after gene therapy. |
| The patients’ leukemic clones had different retroviral insertion sites, but both were near the 5′ end of the LIM domain only LMO-2 transcription factor gene. |
| Both patients have required chemotherapy, and the first received an allogeneic stem cell transplant. |
| Retrovirus vectors, chosen for their ability to effect permanent correction by integrating into the host chromosomal DNA, do not permit control of insertion site. |
| Possible harmful interactions between endogenous host factors (age, disease-specific effects, other inherited susceptibility genes, infections) and host genes whose expression is altered by nearby retroviral insertion (transcription factors, such as LMO-2) can combine to produce leukemia. |
| *Data from Kohn et al23; A. Fischer and M. Cavazzana-Calvo, personal communication in oral presentations, 2003 |
| Advantages |
| Complications and late failures after allogeneic transplantation mandate a search for better treatment. |
| Ex-vivo transduction: hematopoietic cells can be removed and enriched, cultured with retrovirus and then reinfused intravenously, where they will home to the bone marrow. |
| Disease gene product, the common gamma chain of cytokine receptors is widely expressed in blood lineages; overexpression apparently not harmful. |
| The problem of immune elimination of the corrected cells should not arise because of the immunodeficiency phenotype. |
| There is an in vivo selective advantage for lymphoid cells expressing the normal common gamma chain protein. |
| Animal models and human trials have been successful in restoring immunocompetence. |
| Of 10 infants given gene therapy as primary treatment in France by the group of Fischer et al, 9 had restoration of T-cell function.31,32 |
| Specific antibody production was restored in most. |
| A dose of > 1 million corrected CD34+ cells/kg was established as required to achieve full reconstitution. |
| Disadvantages* |
| Serious adverse events occurred in 2/10 infants: development of clonal leukemic proliferations of transduced cells. |
| Both were the youngest infants at treatment (1 and 3 months). |
| Clonal proliferations were detected about 2½ years after gene therapy. |
| The patients’ leukemic clones had different retroviral insertion sites, but both were near the 5′ end of the LIM domain only LMO-2 transcription factor gene. |
| Both patients have required chemotherapy, and the first received an allogeneic stem cell transplant. |
| Retrovirus vectors, chosen for their ability to effect permanent correction by integrating into the host chromosomal DNA, do not permit control of insertion site. |
| Possible harmful interactions between endogenous host factors (age, disease-specific effects, other inherited susceptibility genes, infections) and host genes whose expression is altered by nearby retroviral insertion (transcription factors, such as LMO-2) can combine to produce leukemia. |
Schematic representation of activating and inhibitory immune receptors.
The prototype activating receptor is composed of a glycoprotein subunit containing an extracellular domain (often with an immunoglobulin-like or lectin-like structure) that is responsible for ligand binding. The ligand-binding receptor lacks intrinsic signaling activity, but pairs with an adapter protein possessing one or more ITAM in the cytoplasmic domain. The immunoreceptor tyrosine-based activation motifs (ITAM)-bearing adaptor protein, such as DAP12, FcεRIγ or CD3ζ are expressed on the cell surface as disulfide-bonded dimers; DAP12 is a homodimer, whereas FcεRIγ or CD3ζ, may form homodimers or heterodimers with each other. After ligand binding, tyrosine phosphorylation of the ITAM causes recruitment and activation of Syk or ZAP70 tyrosine kinases. The receptor and signaling adapter associate via interactions in their transmembranes, typically by a salt bridge formed by oppositely charged amino acids. The prototype inhibitory receptor has a ligand-binding extracellular domain and one or more immunoreceptor tyrosine-based inhibition motifs (ITIM) in the cytoplasmic region of the polypeptide. Binding of ligand causes tyrosine phosphorylation of the ITIM, resulting in recruitment and activation of a tyrosine phosphatase (SHP-1 or SHP-2) or a lipid phosphatase (SHIP-1 or SHIP-2).
Schematic representation of activating and inhibitory immune receptors.
The prototype activating receptor is composed of a glycoprotein subunit containing an extracellular domain (often with an immunoglobulin-like or lectin-like structure) that is responsible for ligand binding. The ligand-binding receptor lacks intrinsic signaling activity, but pairs with an adapter protein possessing one or more ITAM in the cytoplasmic domain. The immunoreceptor tyrosine-based activation motifs (ITAM)-bearing adaptor protein, such as DAP12, FcεRIγ or CD3ζ are expressed on the cell surface as disulfide-bonded dimers; DAP12 is a homodimer, whereas FcεRIγ or CD3ζ, may form homodimers or heterodimers with each other. After ligand binding, tyrosine phosphorylation of the ITAM causes recruitment and activation of Syk or ZAP70 tyrosine kinases. The receptor and signaling adapter associate via interactions in their transmembranes, typically by a salt bridge formed by oppositely charged amino acids. The prototype inhibitory receptor has a ligand-binding extracellular domain and one or more immunoreceptor tyrosine-based inhibition motifs (ITIM) in the cytoplasmic region of the polypeptide. Binding of ligand causes tyrosine phosphorylation of the ITIM, resulting in recruitment and activation of a tyrosine phosphatase (SHP-1 or SHP-2) or a lipid phosphatase (SHIP-1 or SHIP-2).