Abstract
Recent technological advances in genomics have led to the discovery of new somatic mutations and have brought deeper insights into clonal diversity. This discovery has changed not only the understanding of disease mechanisms but also the diagnostics and clinical management of bone marrow failure. The clinical applications of genomics include enhancement of current prognostic schemas, prediction of sensitivity or refractoriness to treatments, and conceptualization and selective application of targeted therapies. However, beyond these traditional clinical aspects, complex hierarchical clonal architecture has been uncovered and linked to the current concepts of leukemogenesis and stem cell biology. Detection of clonal mutations, otherwise typical of myelodysplastic syndrome, in the course of aplastic anemia (AA) and paroxysmal nocturnal hemoglobinuria has led to new pathogenic concepts in these conditions and created a new link between AA and its clonal complications, such as post-AA and paroxysmal nocturnal hemoglobinuria. Distinctions among founder vs subclonal mutations, types of clonal evolution (linear or branching), and biological features of individual mutations (sweeping, persistent, or vanishing) will allow for better predictions of the biologic impact they impart in individual cases. As clonal markers, mutations can be used for monitoring clonal dynamics of the stem cell compartment during physiologic aging, disease processes, and leukemic evolution.
Learning Objectives
Understand aspects of clonality, clonal hierarchy, and dynamics in current concepts of bone marrow failure and physiologic hematopoiesis
Establish a connection between somatic mutations and pathogenesis of aplastic anemia
Acquire the ability to infer clinical information and improve interpretation skills of deep next-generation sequencing in bone marrow failure syndromes
Molecular diagnostics in bone marrow failure
Used as a discovery tool, next-generation sequencing (NGS) has led to the identification of somatic and germ line mutations in a large number of genes. These genetic alterations constitute the key pathogenic events in myeloid neoplasias (eg, myelodysplastic syndromes [MDSs]) and in classically nonclonal bone marrow failure syndromes (eg, aplastic anemia [AA]). The most obvious implications of somatic mutations for clinical care are their diagnostic utility for a more precise and objective classification of disease entities or prognostication and targeted drug development and personalized therapy.
Somatic mutational events can serve as clonal markers to assess clonality of hematopoiesis in a more quantitative fashion than was possible with the use of classical X chromosome inactivation studies. Consequently, in addition to a nosologic definition of mutational spectra according to leukemia subentities, NGS technologies have allowed for newer insights into the dynamics of hematopoietic stem cell (HSC) compartment and clonal evolution involving progressive steps of leukemogenesis. An advantage of whole-exome sequencing is comprehensive analysis of mutational diversity and load, including passenger and driver mutations. Quantitatively, along with somatic chromosomal aberrations, the total volume of acquired mutations (mutational burden) may serve as a measure of genomic damage. Deep NGS, especially when applied serially, enables reconstruction of the clonal architecture for individual cases and clonal dynamics (eg, during disease initiation, progression changes resulting from therapies as in minimal residual disease monitoring).1-5
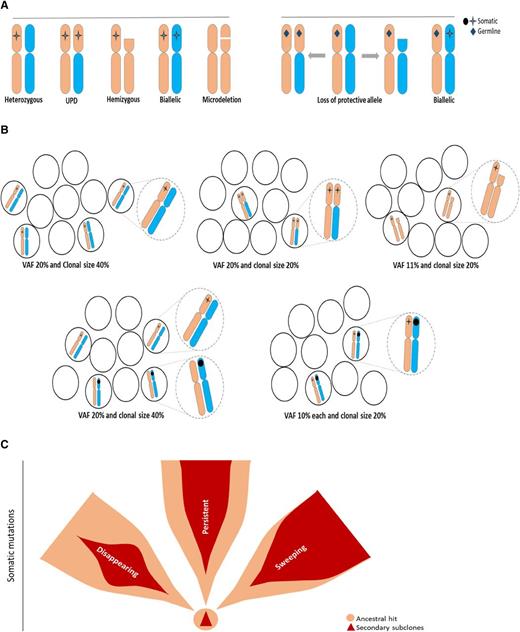
HSC, clonal markers, and principles of clonal architecture
Monitoring of cytogenetic abnormalities often present in MDS has improved our understanding of HSC kinetics in this disease. However, somatic mutational markers can be better quantitated by using the variant allelic frequency (VAF) concept and, thus, can allow for improved precision and resolution (Figure 1). Obvious limitations of the VAF method exist, with its inherent inability to distinguish small clones with copy number-neutral loss of heterozygosity and hemizygous deletions from larger heterozygous clones (Figure 1A-B). Interaction between chromosomal lesions and somatic or germ line events result in a large spectrum of configurations that modify the phenotypic impact of individual lesions (Figure 1A). With regard to theoretical consequences of somatic or germ line events on various configurations of chromosomal and mutational lesions or, for instance, for TP53 mutations, the presence of clones with a biallelic inactivation may be more consequential than the presence of larger heterozygous mutant clones. Such a clonal outgrowth is initiated by specific founder/ancestral events followed by a number of secondary events that lead to either sweeping or less-progressive subclonal expansion (Figure 1C), with linear or branching secondary events contributing to often complex clonal architecture (Figure 2A-B). The analytic imprecision in differentiating mutations in linear configuration from the subclonal mosaicism of concomitant small clones also is important to understand (Figures 1B and 2C).
Configurations of somatic and germ line mutations and VAF concept. (A) Possible configurations of somatic mutations (left) or germ line (right) and chromosomal abnormalities. (B) Impact of clonal configuration on VAF vs calculated clonal burden, including impact of homozygous, hemizygous, or heterozygous mutations (dotted circles show magnified representations of the illustrations in each setting as described). (C) Somatic ancestral vs secondary hits and potential clinical behavior of secondary clones as persistent, disappearing, and sweeping. Ancestral hits are the only somatic events that can occur as single hits. Secondary hits can be present only in association with the ancestral hit. Secondary mutations create a scenario in which only very few single mutant cells exist. Sweeping clones/mutations outgrow single mutant clones, and thus, their VAF may be very similar to that of founder mutations.
Configurations of somatic and germ line mutations and VAF concept. (A) Possible configurations of somatic mutations (left) or germ line (right) and chromosomal abnormalities. (B) Impact of clonal configuration on VAF vs calculated clonal burden, including impact of homozygous, hemizygous, or heterozygous mutations (dotted circles show magnified representations of the illustrations in each setting as described). (C) Somatic ancestral vs secondary hits and potential clinical behavior of secondary clones as persistent, disappearing, and sweeping. Ancestral hits are the only somatic events that can occur as single hits. Secondary hits can be present only in association with the ancestral hit. Secondary mutations create a scenario in which only very few single mutant cells exist. Sweeping clones/mutations outgrow single mutant clones, and thus, their VAF may be very similar to that of founder mutations.
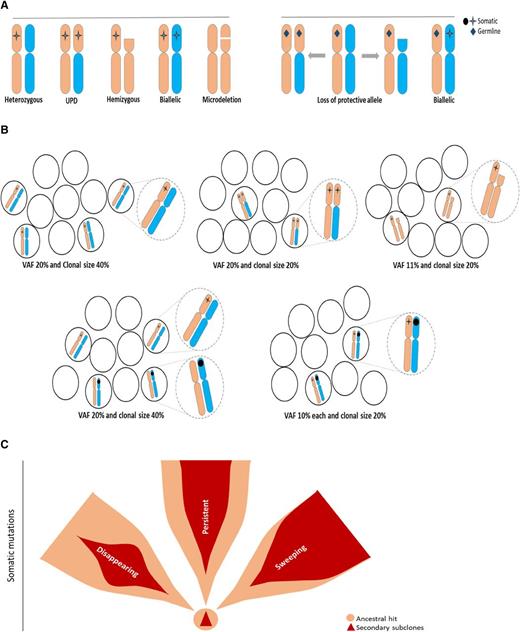
Clonal architecture and types of mutational events, including ancestral and subclonal mutations. Somatic ancestral events are the only clones (A-B) followed by secondary events. (A) Linear evolution with a secondary subclone. (B) Branching evolution with 2 subclones. (C) Ancestral event occurs in germ line mutations and is nonclonal. The somatic hits are subclonal. (D) Codominant events occurring in close succession and thereby not distinguishable by VAF as a primary vs secondary hit.
Clonal architecture and types of mutational events, including ancestral and subclonal mutations. Somatic ancestral events are the only clones (A-B) followed by secondary events. (A) Linear evolution with a secondary subclone. (B) Branching evolution with 2 subclones. (C) Ancestral event occurs in germ line mutations and is nonclonal. The somatic hits are subclonal. (D) Codominant events occurring in close succession and thereby not distinguishable by VAF as a primary vs secondary hit.
Somatic mutations and clonal architecture in MDS
Theoretically, molecular subclassification that is based on the initial hits, such as for core-binding factor acute myeloid leukemia (AML), should also be possible in MDS. However, current characterization of mutational spectra as a result of the diversity and apparent lack of subtype specificity6-8 does not allow for precise mutation-based classifications except for a few inherited leukemia syndromes (germ line RUNX1, DDX41, and CEBPA)9-11 and some invariant balanced translocations [eg, t(8;21) AML, irrespective of morphology] with diagnostic molecular abnormalities.12 Notwithstanding these problems, with diagnostic application of mutational spectra in MDS to validate the morphology as a gold standard, the earliest (ancestral) hits possibly allow for molecular distinction of MDS types. Typical, albeit not pathognomonic, spectra or individual mutations have been described for some phenotypes (eg, chronic myelomonocytic leukemia, refractory anemia with ring sideroblasts with marked thrombocytosis, refractory anemia with ring sideroblasts, paroxysmal nocturnal hemoglobinuria [PNH]).13-15 Ancestral events can be classified as of germ line origin and thus, by definition, as nonclonal (as seen in inherited leukemia traits), or they can be somatic. By using the VAF concept, somatic ancestral alteration can be distinguished by hierarchical ranking as the largest clone (Figure 2C). Because of quick succession of subsequent sweeping events, the clonal succession often cannot be resolved, resulting in ≥ 2 alterations that appear as codominant events (Figure 2D).
In MDS, mutations in some genes are categorically subclonal, whereas others can be either ancestral or subclonal. Typical AML founder lesions, such as those seen in core-binding factor AML or chronic myeloid leukemia, often are self-sufficient. However, others often can persist asymptomatically until secondary hits occur, leading to progression. Some of the initial events likely are ubiquitous and do not predetermine disease specificity, whereas certain secondary subclonal events may shape the phenotype or evolution. In contrast, some founder events may be associated with a specific morphology and/or speed of subsequent progression. For instance, subclonal TET2 occurring as a second hit after ancestral TET2 mutation will lead to a myeloproliferative phenotype. Similarly, initial hits of some genes, such as CEBPA, TP53, and EVI1, can predispose to inactivation of the remaining allele by additional mutations in the contralateral allele through somatic uniparental disomy with duplication of the mutant allele or by deletion events.16-19 Of note, biallelic hits may be either as germ line/somatic or as 2 distinct somatic events.
Generally, MDS can be initiated by preclinical clonal hematopoiesis now identifiable by the presence of clonal mutations in certain genes, including DNMT3A, TET2, SF3B1, and others.20,21 This condition, which often is asymptomatic, is now referred to as clonal hematopoiesis of indeterminate potential (CHIP). Ultimately, the presence of CHIP may have unfavorable implications. CHIP prevalence increases with age, although some genes, such as TET2, are disproportionately overrepresented among the elderly, and noncanonical DNMT3A mutations constitute the bulk of CHIP lesions. Although all founder lesions appear to be promoted by aging factors, some are more permissive/virulent and lead to progression without a period of asymptomatic CHIP. For some of the lesions, their position within the clonal hierarchy may be of importance to their impact on prognosis, whereas for others, their position within the clonal hierarchy is not consequential.
To date, available diagnostic and prognostic information is based on several large studies (Table 1).6-8,22-25 Analogous to assignment of risk for specific cytogenetic defects, clinically relevant prognostic information already has been assigned to various mutations, such as TP53, the RAS gene family, SETBP1, and DNMT3A, in terms of survival outcomes and results of specific therapies.26-29 Of note, in genes where mutations are not totally canonical, individual mutations may have a distinct prognostic impact (eg, as seen with DNMT3AR882 vs truncating DNMT3A mutations).30 The presence of specific mutations increasingly will dictate possible therapeutic choices (Table 2). Although the frequencies of certain mutations will vary among disease subentities, in most instances, they do not secure the diagnosis in an individual patient. Thus, attempts to identify molecular patterns that would completely match the morphologic gold standard have not yet been successful. However, future classification schemes may rely more on mutational/molecular than on morphologic criteria.
Somatic mutations and clonal architecture in AA and PNH
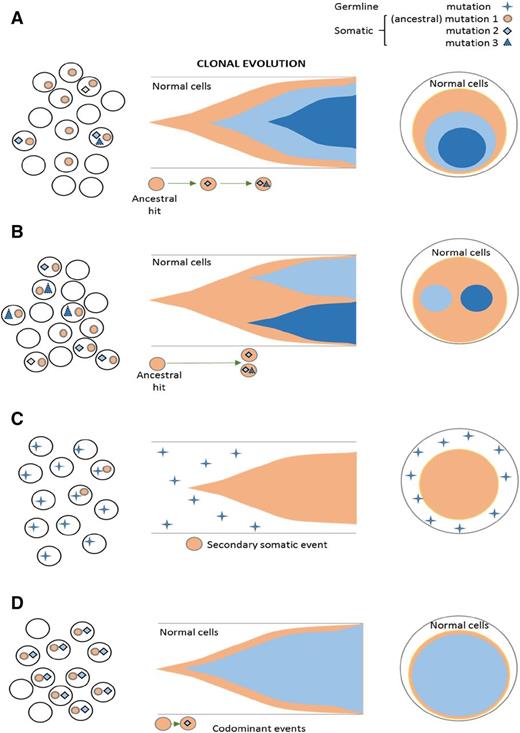
Theoretically, contraction of the normal HSC pool can lead to oligoclonality (Figure 3A). For instance, in AA, clonal chromosomal abnormalities can appear transiently because they can be diluted out by reexpansion of the normal HSC pool during recovery of normal hematopoiesis.31 A similar observation has been made with deep NGS-based mutational analysis, often showing intermittent appearance of clones characterized by somatic mutational events.22 The fate of the corresponding clone possibly depends on the type of mutation (gene affected). Some of the mutant clones found at the initial presentation can be outgrown by recovering normal clones (Figure 3B), or conversely, they can be selected for and expanded, resulting in MDS or PNH (Figure 3A). Alternatively, some mutations, if occurring in improper sequence, and thus molecular ranking context within clonal hierarchy (secondary hits without founder background) cannot persist, especially when normal HSCs have recovered. In either case, clonality as a result of normal HSC contraction (eg, in AA, in aging hematopoiesis) can be contrasted with the true malignant expansion owing to intrinsic growth advantage or external selection pressure (Figure 3A,D). The existence of clonal mutations in AA has been inferred from the presence of PNH clones identified by sensitive PNH flow cytometry.32
HSC compartment and modes of clonality of hematopoiesis in bone marrow failure (A) Clonality or oligoclonality can be due to HSC depletion (eg, in some cases of AA) or a true subclonal expansion. (B) Transient clonal evolution in AA. (C) Theories about secondary MDS evolution in the context of AA. Mutant (clonal) HSC can already exist at presentation with subsequent selection (top), or the clonogenic hits occur in the course of disease (eg, as a result of endogenous/immune-mediated mutagenesis [bottom]). (D) Progressive linear acquisition of mutations as seen in typical MDS. CTL, cytotoxic T lymphocyte; GF, growth factor.
HSC compartment and modes of clonality of hematopoiesis in bone marrow failure (A) Clonality or oligoclonality can be due to HSC depletion (eg, in some cases of AA) or a true subclonal expansion. (B) Transient clonal evolution in AA. (C) Theories about secondary MDS evolution in the context of AA. Mutant (clonal) HSC can already exist at presentation with subsequent selection (top), or the clonogenic hits occur in the course of disease (eg, as a result of endogenous/immune-mediated mutagenesis [bottom]). (D) Progressive linear acquisition of mutations as seen in typical MDS. CTL, cytotoxic T lymphocyte; GF, growth factor.
Deep sequencing studies revealed that PNH is not strictly monogenic in the sense that multiple clones with PIGA mutations may coexist. In addition, some PNH clones display a more complex subclonal structure that involves not only PIGA mutations but also mutations in other myeloid genes typical of MDS.25 Some aspects of clonal evolution are not completely explained by the extrinsic immune selection theory of PNH. For instance, even after successful immunosuppression, clonal expansion continues in some patients, whereas in others, PNH clones remain stable. The presence of additional clonal or subclonal mutations serving extrinsic factors has been predicted to explain some of the clinical heterogeneity in PNH and is now confirmed by comprehensive sequencing studies.25 In general, PIGA mutations constituting an ancestral event and subclonal branching mutation of various genes, similar to those encountered in MDS, have been observed. However, occasional PIGA mutations are also subclonal, trailing the ancestral non-PNH hits similar to those observed in AA. Finally, in some instances, the PIGA mutant clone coexists with clones characterized by other mutations in the form of clonal mosaicism.
The evolution of MDS in AA patients is a serious complication with major therapeutic implications, with a rate of evolution ∼10% to 20% in 10 years.19,31,33 Most of the secondary MDSs evolved from AA are characterized by acquisition of monosomy 7, frequently as a singular chromosomal aberration with a poor prognosis.33 Single-nucleotide polymorphism array karyotyping may be particularly helpful if metaphases have not been obtained as a result of paucicellular aspirate.19 For instance, the occurrence of microdeletions or UPD6p involving portions of the HLA locus (leading to loss of one of the HLA alleles presenting immunodominant antigens) has been found,19,34,35 likely evolving as an escape phenomenon from immune selection pressure. Similarly, inactivating mutations in the HLA locus may be selected for during immune attack; this has been shown for HLA-B*40:02. Various somatic alterations, such as frameshift, nonsense, and splice site mutations, were found in both patients with and without UPD6p context, suggesting that immune selection, perhaps analogous to PNH, leads to evolution of clones with inactivating molecular lesions in HLA loci.36
Clonal evolution and cytogenetic abnormalities may be selected on the basis of other underlying mechanisms. For instance, some of the cases with −7/del(7q) correspond to escape processes resulting from somatic deletions of gain-of-function SAMD9 and SAMD9L mutant variants conveying bone marrow failure.37-39 Other mechanisms also may be involved as indicated by a relatively high frequency of −7/del(7q) in patients with GATA2 mutations.40 Clinically, no predictive factors have been identified, but most of the patients who evolve show either primarily refractory disease or less-than-optimal recovery after immunosuppression.33 To date, no recurrent hemizygous hits have been identified on the retained allele in post-AA MDS with −7/del(7q), but various mutations often encountered in MDS can be found, including SETBP1, CBL, RUNX1, and others.22,41,42 Systematic application of whole-exome sequencing or targeted deep sequencing to AA patients in a cross-sectional and serial setting has led to the discovery that clones characterized by somatic mutations are present in a proportion of patients.22,34,42,43 These mutations, in addition to those in PIGA, affect many other typical myeloid genes, including DNMT3A, CBL, SETBP1, TET2, ASXL1, BCOR, and BCORL. Whether increased frequency of certain events has pathophysiologic meaning (eg, milieu favoring certain types of mutations) or reflect bias stemming from a much smaller cohort of patients and from a number of studies performed to date in AA compared with MDS is unclear. Overall, most of these mutations are present only in tiny clones as assessed by deep NGS. Serial studies have revealed that some of these clones do not persist but can expand and disappear, which is particularly true for BCOR, BCORL, and DNMT3A mutant clones, suggesting a limited recruitment choice in a contracted stem cell pool (Figure 3A). Moreover, the presence of a clonal mutation by itself does not predict future evolution of clinical MDS; rather, only certain hits with a sufficient virulence are more likely to be selected and to successfully maintain clonal outgrowth. Subsequent secondary hits may further accelerate this process. In contrast, less-permissive mutations may be either eliminated by the immune system or, if not supported, by secondary lesions unable to compete against the recovering normal HSCs. Of note, some of the clones identified in AA may correspond to the normally occurring clonal outgrowth present in asymptomatic fashion, with increasing frequency in elderly individuals in the form of CHIP. Therefore, their detection would equate to the discovery of coexistent CHIP in patients with AA (Figure 3C).
Deep NGS performed to trace clonal origin back to the presentation of AA demonstrated that subclonal expansions already exist at presentation in a proportion of AA patients.41 This discovery highlights the possibility rather than reflects the true frequency of this phenomenon because of sensitivity limitations of deep NGS. However, some of the hits (eg, RUNX, SETBP1) already present at the beginning of the process may give rise to later secondary MDS and suggests that del7 is not the ancestral defect but, rather, is secondary to initial founder mutations.
One important scientific implication of clonal mutations found in AA pertains to the dynamics of the HSC compartment and their corroboration with theories about the origins of idiopathic AA. One could envision how mutational events could trigger immune tumor surveillance response, which could cross-react with and damage HSCs, resulting in AA (Figure 3); eliminate the pathologic clones; or indeed lead to selection of the most fit mutant clones that escape immune surveillance and lead to secondary MDS.
In conclusion, from a clinical perspective, firm prognostic and diagnostic definitions are not yet possible. Some mutational events clearly are compatible with normal hematopoietic function and spontaneously retract, whereas others clearly are pathogenic and will show prognostic impact and be highly predictive of the risk of evolution. Future studies could provide this distinction. Currently, the general presence of mutational events at the onset of disease precludes response to immunosuppressive therapies, but some of the mutations may not be favorable.
Correspondence
Jaroslaw P. Maciejewski, Department of Translational Hematology and Oncology Research, Taussig Cancer Institute, Cleveland Clinic, 9500 Euclid Ave, Desk R40, Cleveland, OH 44195; e-mail: maciejj@ccf.org.
References
Competing Interests
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Author notes
Off-label drug use: None disclosed.


![Figure 3. HSC compartment and modes of clonality of hematopoiesis in bone marrow failure (A) Clonality or oligoclonality can be due to HSC depletion (eg, in some cases of AA) or a true subclonal expansion. (B) Transient clonal evolution in AA. (C) Theories about secondary MDS evolution in the context of AA. Mutant (clonal) HSC can already exist at presentation with subsequent selection (top), or the clonogenic hits occur in the course of disease (eg, as a result of endogenous/immune-mediated mutagenesis [bottom]). (D) Progressive linear acquisition of mutations as seen in typical MDS. CTL, cytotoxic T lymphocyte; GF, growth factor.](https://ash.silverchair-cdn.com/ash/content_public/journal/hematology/2017/1/10.1182_asheducation-2017.1.66/4/m_hem00008f3.jpeg?Expires=1765943741&Signature=SXx91jVgToN91Vrb0pxEy1CVz1e162fsbWtqyX1qpXcva5QxUfE1AOMQhT9xa8CzivnTu28s3VXKN2ynvuKula5r4Si-TDrWtdIwwGdZ~Pj4qrKzkjop71T-wsmbfw5MnFLLvhZl4XJdIR9N-5b1Unx6fMReGEmnqC7tp~8Ser-3ZpymFhixF36Qa3dz6-JapLoj2Pn8pEgZ1ob1vfTdpJ3Ftp6cYBFlexdknz31OJEiEoDHJIPnIsGYnxza8VTm7jjGmWy3bW4KU7AoFuKwcX5ysWpjK7JdllV24UwfHtHVh4rT5FJpgpZ4gL57kx-G0MluP3Ed~IjcYFehdtNuoQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. HSC compartment and modes of clonality of hematopoiesis in bone marrow failure (A) Clonality or oligoclonality can be due to HSC depletion (eg, in some cases of AA) or a true subclonal expansion. (B) Transient clonal evolution in AA. (C) Theories about secondary MDS evolution in the context of AA. Mutant (clonal) HSC can already exist at presentation with subsequent selection (top), or the clonogenic hits occur in the course of disease (eg, as a result of endogenous/immune-mediated mutagenesis [bottom]). (D) Progressive linear acquisition of mutations as seen in typical MDS. CTL, cytotoxic T lymphocyte; GF, growth factor.](https://ash.silverchair-cdn.com/ash/content_public/journal/hematology/2017/1/10.1182_asheducation-2017.1.66/4/m_hem00008f3.jpeg?Expires=1765943742&Signature=G4N-5WlVtdrFLsaAKubWT6DP6pmEYcLyp9JWEguurLoxDhK4Tt-RtwdBHT3nNmMwK~-l~K9iGHHv3OuLJ4A~-XCKabduxV7pgW~p2fbWBWlEWZdkQvzymVJLdss9N6WztP9aXpV3UTfBA3iZB2csvNQb8Rj0nwY7jSIxi9tlkh2ymXcQOUJL5JoGOZh3Okj5jKz8~oyQFeFkns60-TJLtwwHUoLrwZ5aw16PTp8iY1zq~oCuiT3t53RzHkzYcokYWlgy64KDVv~0-WqR83EAcR3VirZtBT3jRWHIrDb4xKS7Bs0u0gtbWjiI5P-UUaTYKgirKetNxWqluk~lSxXSRQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)