Abstract
Diffuse large B-cell lymphomas (DLBCLs), the most common lymphoid malignancies, are clinically and genetically heterogeneous disorders. Although DLBCL is a chemo-responsive tumor, many patients will not be cured with conventional empiric treatment regimens. Gene expression profiles, analyses of specific genetic abnormalities and functional assays have been used to develop comprehensive molecular signatures of tumors that share similar features and rely upon common survival pathways. These studies are leading to the identification of subtype-specific rational therapeutic targets and associated inhibitors for clinical investigation.
Clinical Heterogeneity of Diffuse Large B-Cell Lymphoma
Diffuse large B-cell lymphomas (DLBCLs) are the most common lymphoid malignancies in adults, comprising almost 40% of all lymphomas.1,2 Although more than 60% of patients with DLBCL are cured with empiric combination chemotherapy and rituximab, the remainder relapse and many ultimately die of their disease.2
Genetic Heterogeneity in DLBCL
The majority of DLBCLs are thought to arise from normal antigen-driven B cells that are undergoing clonal expansion in the germinal centers (GCs) of peripheral lymphoid organs.3 These GC B cells have completed productive rearrangement of their immunoglobulin genes via V(D)J recombination; in the GC, these cells undergo further class-switch recombination and somatic hypermutation (SHM). This process, which serves as a marker for the GC stage of differentiation, requires double-strand DNA breaks (DSBs) and creates an optimal setting for errors in DNA repair. Up to 50% of DLBCLs harbor translocations involving immunoglobulin loci and cellular oncogenes. Since these tumors arise from GC B cells undergoing IgH class-switch recombination, somatic mutation and possible further V(D)J rearrangement, translocations involving immunoglobulin loci may be mediated by errors in these processes. The most common chromosomal translocations in DLBCLs bring genes including BCL6, BCL2 and cMYC under the inappropriate control of an immunoglobulin regulatory element (Table 1 ). In additional DLBCLs, aberrant SHM of genes, including BCL6, has been described (Table 1 ).4 Another recurrent genetic lesion is the deletion of the FAS death domain, which limits apoptosis via the extrinsic pathway (Table 1 ).5,6 Inactivating mutations of p53 have also been described in a small number of DLBCLs (Table 1 ). However, many DLBCLs lack the aforementioned genetic abnormalities, suggesting that additional pathogenetic mechanisms remain to be defined.
Morphologic Heterogeneity in DLBCL
In DLBCL, the malignant B cells are large transformed lymphocytes that diffusely efface the normal architecture of involved nodal or extranodal sites. Specific variants of LBCL, including primary mediastinal LBCL (MLBCL) and T-cell/histiocyte-rich BCL (T/HRBCL), have been defined on the basis of clinical presentation and/or specific morphologic features.7
MLBCL
Unlike DLBCL, which commonly arises in elderly patients of both sexes, MLBCL typically presents in younger women.8,9 Patients with MLBCL have bulky mediastinal masses with frequent invasion of the adjacent structures. These tumors also exhibit several characteristic genetic abnormalities such as gains of chromosomes 9p and 2p which include the JAK2 (9p24) and REL (2p16) loci.8,10
T/HRBCL
T/HRBCLs typically have a dense infiltrate of polyclonal T cells surrounding smaller numbers of malignant B cells with morphologic and immunophenotypic features of DLBCLs.2,11–15 These tumors occur in somewhat younger patients, often involve the liver, spleen and bone marrow, and exhibit fewer genetic abnormalities.14
Analysis of DLBCL Heterogeneity
The striking clinical, genetic and morphologic heterogeneity in DLBCL and its variants suggests that additional subtypes and biologically relevant substructures remain to be defined. To understand the bases of clinical and molecular heterogeneity in DLBCL, it would be useful to have comprehensive molecular signatures of tumors that share similar features.11,16 In addition to highlighting potential pathogenetic mechanisms, these signatures might identify promising subtype-specific targets for therapeutic intervention. It is now possible to obtain signatures of biologically relevant DLBCL subtypes using gene expression profiling (GEP). To date, transcriptional profiling of DLBCLs has been used to: (1) identify features associated with unfavorable responses to empiric combination chemotherapy; (2) highlight similarities between certain tumors and subsets of normal B cells; and (3) define robust and highly reproducible DLBCL subtypes with comprehensive transcriptional signatures.2
Outcome
In earlier profiling studies, the molecular signatures of DLBCLs with different responses to standard chemotherapy were directly examined.17 Signatures predictive of outcome (cured vs fatal/refractory disease) were identified, which included genes involved in B-cell receptor signaling, regulation of apoptosis, and serine/threonine phosphorylation, among others.17 Of the genes and pathways associated with poor responses to current regimens, two have already been directly examined and targeted for possible therapeutic intervention (PKCβ and the cyclic AMP-specific phosphodiesterase PDE4B).18–20 A multicenter phase 2 study of a potent inhibitor of PKCβ, enzastaurin, was conducted in patients with recurring or refractory DLBCL.19 In this study, treatment with the single-agent oral PKCβ inhibitor was associated with freedom from progression (FFP) for more than 2 cycles in 22% of patients. Of particular interest, 4 patients continue to be progression free for 20 to more than 50 months following study entry.19
Cell-of-Origin
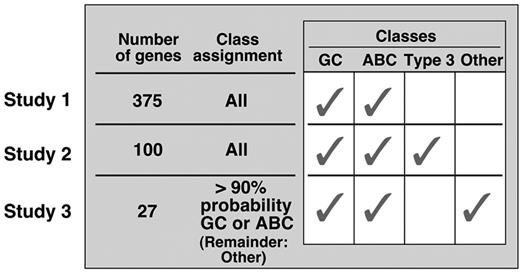
A series of molecular models have been described that relate subsets of DLBCL to normal B cells at different stages of development (Figure 1 ).2 Initially, two groups of DLBCLs were identified—GC-like and activated B-cell–like (ABC)—based on similarities in expression of 375 genes in the tumors and normal GC B cells or in vitro–activated peripheral blood B cells.21 In this study, DLBCLs with features common to normal GC B cells responded more favorably to standard empiric chemotherapy.21 The initial cell-of-origin (COO) signature was revised to include only 100 genes that delineated GC-and ABC-like DLBCLs; a third group of tumors did not fit into either category (Type 3).22 Thereafter, using the same dataset, the COO signature was further refined and limited to only 27 genes, which delineated GC- and ABC-like tumors; a third unspecified category (“Other”) was also described.23 Although the 100- and 27-gene models largely identified the same tumors as either GC-or ABC-like, there was poor agreement on the third default category (Type 3/Other), which includes up to 40% of DLBCLs in other recent series.2,11,23 In more recent clinical trials of rituximab-containing combination chemotherapy, the prognostic significance of COO is no longer apparent.24
Distinct DLBCL Subtypes: MLBCL versus DLBCL
Two groups of investigators have compared the gene expression profiles of newly diagnosed MLBCL and DLBCL to identify unique molecular features of MLBCL.10,25 Primary MLBCLs expressed low levels of B-cell receptor (BCR)–signaling pathway components and exhibited a distinctive cytokine signature, features that were strikingly similar to those of classical HL (cHL).10,25 The resemblance between the transcription profiles of MLBCL and cHL was of particular interest because these diseases have similar clinical presentations and share specific genetic lesions such as gains of chromosomes 9p and 2p.10 In both MLBCL and cHL, the identification of chromosome 9p gains has prompted further analyses of JAK2 signaling and additional mechanisms of deregulating this pathway, including deletions or inactivating mutations of SOCS1.26
The known role of NF-κB activation in cHL and the increased expression of certain NF-κB target genes in primary MLBCLs prompted further investigation of NF-κB survival pathway in MLBCL using immunohistochemical methods and functional assays.10,27 There was near-uniform nuclear localization of the cREL NF-κB subunit in primary MLBCLs, indicating that the NF-κB survival pathway was constitutively active in these tumors.10,27 In a multi-institutional follow-up study, investigators demonstrated the utility of subcellular (nuclear) cREL localization and expression of an additional NF-κB target gene, TRAF, in diagnosing MLBCL.28 In addition, functional assays confirmed that MLBCL cell lines exhibited increased constitutive NF-κB activity and relied upon the NF-κB pathway for survival.27,29 Therefore, the MBLCL transcriptional profile has already been translated into a functionally relevant immunohistochemical signature in this disease. As specific NF-κB inhibitors become available for clinical trials, MLBCL should be rapidly targeted for further evaluation.
DLBCL Consensus Clusters
Our investigative group believed that there were important differences between normal and malignant B cells and asked whether previously undefined subtypes of DLBCL might have other distinguishing characteristics. For these reasons, we analyzed the transcriptional profiles of an additional large series of newly diagnosed DLBCLs with three different clustering algorithms. The top 5% of genes with the highest reproducibility across duplicate samples and the largest variation across tumors were used.11 In these studies, we used a resampling-based method (consensus clustering) that automatically selects the most stable number of tumor groups (clusters) with each algorithm.11 As previously reported, three robust clusters of DLBCLs were identified.11 The same approach was also applied to a large independent DLBCL dataset, and the same three groups of tumors were seen.11 Of note, the consensus clusters identified subsets of DLBCLs that were different from those defined by their developmental signature (COO).11 In the two large independent datasets, both types of DLBCL substructure (consensus clusters and COO) were readily detectable,11 indicating that the two classification methods captured different aspects of DLBCL heterogeneity.
Analysis of DLBCL consensus clusters
Ox Phos DLBCLs
The Ox Phos cluster showed increased expression of genes regulating mitochondrial function, electron transport, apoptosis, and proteosomes.11 Genetically, these tumors were more likely than others to exhibit genetic lesions affecting the intrinsic and extrinsic apoptotic pathways including t(14;18) and deletion of the FAS death domain.6,11
BCR tumors
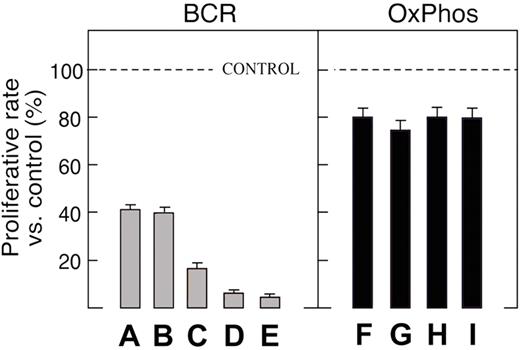
BCR DLBCLs had increased expression of cell-cycle regulatory genes, DNA repair genes, components of the B-cell receptor signaling cascade, and numerous B-cell–specific transcription factors, including BCL6.11 In addition, translocations involving BCL6 [t(3q27;…)] were significantly more common in BCR DLBCLs.6,11 Since DLBCLs dependent on BCL6-regulated pathways should exhibit differential regulation of BCL6 target genes, we recently used chromatin immunoprecipitation (ChIP)-on-chip to identify direct BCL6 target genes, which included modulators of transcription, chromatin structure, protein ubiquitylation, cell cycle, and DNA damage responses.30 These BCL6 target genes were clearly differentially regulated in BCR DLBCLs.30 In a panel of DLBCL cell lines analyzed by expression arrays and classified according to their gene expression profiles, only BCR tumors were highly sensitive to the BCL6 peptide inhibitor, BPI (Figure 2 ).30 Of note, the consensus cluster designation (BCR DLBCL) was more effective in predicting BPI sensitivity than baseline BCL6 expression.30 Taken together, these studies suggest that BCR DLBCLs are more reliant upon BCL6 signaling and uniquely sensitive to BCL6 inhibitors. From a clinical standpoint, these data also suggest that patients with BCR DLBCLs may represent the best candidates for therapeutic trials of BCL6 inhibitors. Ongoing studies also indicate that BCR DLBCLs may be particularly reliant upon tonic BCR survival signals and sensitive to targeted pharmacologic inhibition of the BCR pathway.32
HR DLBCLs
Unlike BCR and Ox Phos DLBCLs, HR tumors exhibited a brisk host immune/inflammatory response.11 HR tumors had a prominent T-cell/dendritic cell infiltrate like that seen in the previously described morphologic DLBCL subtype, T/HRBCL.2,11–15 In addition, patients with HR tumors more commonly had the same clinical features as patients with T/HRBCL including younger age, more frequent liver, spleen and bone marrow involvement, and fewer known genetic abnormalities.6,11 These data indicate that DLBCLs defined by transcriptional profiling as HR tumors include those previously designated as T/HRBCLs on the basis of morphologic, clinical and genetic features. Of note, patients with HR DLBCLs do not have more favorable outcomes, prompting speculation that the brisk host immune/inflammatory response is ineffective or inhibited by counter-regulatory measures.2 An alternative hypothesis is that the tumor-infiltrating T cells actually function to promote DLBCL cell growth.33 The availability of precise molecular signatures of HR DLBCLs allows us to further investigate these possibilities.
Future Directions
New insights into the molecular heterogeneity of LBCLs will likely improve our ability to diagnose entities such as T/HRBCL and primary MLBCL. These tumors share characteristics of large B-cell lymphomas and certain HLs (nodular lymphocyte predominant HL [NLPHL] and cHL, respectively), including an increased host inflammatory response (Figure 3; see Color Figures, page 515). Certain features of these “gray zone” lymphomas suggest that these tumors are defined, and possibly driven, by their interactions with the host microenvironment.2 Recent studies are beginning to elucidate the mechanisms used by lymphoma cells to edit host antitumor responses and limit their effectiveness.34 These insights may lead to new targeted approaches to lymphoma immunotherapy.
In addition, the emerging molecular signatures of specific subtypes of LBCL may improve diagnostic accuracy and highlight potential rational treatment targets. For example, the immunohistochemical signature of MLBCL is based, in part, on the constitutive activation of the NF-κB survival pathway in this disease.10,27,28 In addition, consensus cluster–defined “BCR” DLBCLs may be particularly sensitive to targeted inhibition of BCL6 and tonic BCR signaling.30,32 Our increasing understanding of the molecular heterogeneity of LBCL will allow us to identify promising rational treatment targets in specific tumor types and, hopefully, improve their therapy.
Sequential molecular models that relate subsets of DLBCLs to certain normal B cells or leave tumors unassigned (see text).2,21–23
Reprinted with permission from Abramson and Shipp.2
BCR and Ox Phos DLBCL cell lines exhibit differential sensitivity to the BCL6 peptide inhibitor (BPI).
Proliferation of BPI-treated BCR and Ox Phos DLBCL cell lines. Cell lines were treated with 20 μM BPI.
BCR and Ox Phos DLBCL cell lines exhibit differential sensitivity to the BCL6 peptide inhibitor (BPI).
Proliferation of BPI-treated BCR and Ox Phos DLBCL cell lines. Cell lines were treated with 20 μM BPI.
Dana Farber Cancer Institute, Harvard Medical School, Boston, MA