Abstract
Multiple myeloma is a tumor of somatically mutated, isotype-switched plasma cells that accumulate in the bone marrow leading to bone destruction and bone marrow failure. The germinal center processes of somatic hypermutation and switch recombination are implicated in the development of recurrent immunoglobulin gene translocations in 40% of patients. These affect five loci: 11q13, 6p21, 4p16, 16q23 and 20q11, leading to dysregulation of CCND1, CCND2, FGFR3/MMSET, c-MAF and MAFB respectively. The remaining 60% of patients can be divided into four groups based on their expression of CCND1 and CCND2. The largest group (40%) ectopically express CCND1 bi-allelically and have hyperdiploidy with multiple trisomies of chromosomes 3, 5, 7, 9, 11, 15, 19 and 21. The translocation and cyclin D (TC) groups identify patients with different genetics, biology, clinical features, prognosis and response to therapy.
Despite recent advances, multiple myeloma (MM) continues to be an incurable plasma cell (PC) malignancy, with a yearly incidence of 14,000 in the US and a median survival of 3 years. It accounts for nearly 2% of deaths from cancer.1 Often it is preceded by a pre-malignant tumor called mono-clonal gammopathy of undetermined significance (MGUS), which occurs in about 3% of individuals over the age of 50.2,3 It is important to distinguish two kinds of MGUS tumors. IgM MGUS usually has a lymphoplasmacytic phenotype and rarely—if ever—progresses to MM. Non-IgM MGUS tumors (some of which synthesize only a mono-clonal Ig light chain and no heavy chain) mostly have a plasmacytic phenotype and can progress to MM tumors. Non-IgM MGUS and MM both show a marked increased prevalence with age. The prevalence of each tumor is about twofold higher in African Americans than in Caucasians, and there is suggestive evidence for some nonrandom clustering of MM and MGUS within families.4 However, the roles of genetic background and environment remain to be defined.
MM Is a Plasmablast/Plasma Cell Tumor of Post-Germinal Center B Cells
Although there is a similar prevalence of T cell tumors and pre-germinal center (pre-GC) B cell tumors, most B cell tumors involve germinal center (GC) or post-GC B cells that have modified their immunoglobulin (Ig) genes by sequential rounds of somatic hypermutation and antigen selection, and sometimes by IgH switch recombination.5 These two B-cell-specific DNA modification processes, which occur mainly in GC B cells, selectively target V(D)J and switch sequences in Ig genes, respectively, but occasionally they can cause mutations or double strand DNA breaks in or near non-Ig genes, including oncogenes. Post-GC B cells can generate plasmablasts (PB) that have successfully completed somatic hypermutation, antigen selection, and IgH switching.6 Typically, these PB migrate to the bone marrow (BM), where stromal cells facilitate terminal differentiation into long-lived PC. Pre-GC B cells can generate short-lived PC that mostly remain in the primary lymphoid environment. However, MM and non-IgM MGUS are exclusively monoclonal post-GC tumors that have phenotypic features of PB/long-lived PC, and usually are distributed at multiple sites in the BM.7 The variable regions of Ig genes in non-IgM MGUS and MM tumors are extensively mutated, with the pattern of mutations suggesting repeated rounds of somatic hypermutation and antigen selection.8 Although there is a lack of intraclonal heterogeneity of Ig variable region mutations in MM, some non-IgM MGUS tumors have intraclonal heterogeneity, indicating that these MGUS tumors or their precursors retain the capability of somatic hypermutation.9 A critical feature shared by MGUS and MM is an extremely low rate of proliferation, usually with no more than a few percent of cycling cells until later stages of MM.2 Some proliferative tumor cells may have a phenotype that is similar to a PB or a pre-PB that expresses some B cell markers (CD19, CD20, CD45) but not some PC markers (CD138).10 Other proliferative tumor cells may have a more differentiated phenotype that includes expression of CD138.11 Unfortunately, the precise phenotype(s) and location(s) of the proliferating tumor cell remains a contentious issue. However, most tumor cells are non-proliferative. These cells are not fully differentiated, but have a phenotype that typically is similar to normal, terminally differentiated, long-lived BM PC (CD19−CD20−CD45−CD138+). It is tempting to speculate that MM is similar to many kinds of hematopoietic and solid tumors that are thought to be dependent on a small population of stem cells that are capable of indefinite proliferation, but presently there is no convincing evidence for a small fraction of stem cells in MM. Importantly, we do not know whether or not the non-proliferative cells retain the ability to revert to a proliferative phenotype. In any case, the occurrence of tumor cells with different phenotypes is an important consideration in the design and evaluation of therapies.
Stages of Multiple Myeloma
A clonal PC neoplasm must expand to approximately 109 cells before it produces enough Ig to be recognized as a monoclonal Ig (M-Ig) by serum electrophoresis, or as a monoclonal Ig light chain (M-IgL) by urine electrophoresis for the approximately 15% of MM tumors that express only IgL.2 Notably, the relatively recent development and application of a serum free IgL assay has increased the sensitivity and reliability of screening for clonal PC tumors.12 The most significant impact of this procedure includes an enhanced ability to detect M-Ig or M-IgL in primary amyloidosis and nonsecretory multiple myeloma, as well as an improved capability to detect and monitor tumors that express only M-IgL, with preliminary results indicating that a significant fraction of MGUS tumors express only M-IgL. In any case, for MGUS, serum M-Ig is 0.5 to 3 g/dL, and the tumor cells comprise no more than 10% of the mononuclear cells in the BM (Figure 1 ). Depending on the level of M-Ig, 0.6%–3% per year of patients with non-IgM MGUS progress to MM expressing the same M-Ig.3 There are no unequivocal genetic or phenotypic markers that distinguish MGUS from MM tumor cells, so that it is not possible to predict if and when an MGUS tumor will progress to MM. Also, it remains unclear to what extent intrinsic genetic or epigenetic changes in the MGUS tumor cell versus extrinsic changes in non-tumor cells affect progression. Primary amyloidosis is caused by an MGUS tumor (sometimes with such a small number of tumor cells that M-Ig is not detected by serum electrophoresis) that is symptomatic because of pathological deposits of portions of the M-Ig in critical tissues. MM is distinguished from MGUS by having a BM tumor content > 10%. Smoldering MM (SMM), which has a stable BM tumor content of > 10% but no osteolytic lesions or other complications of malignant MM, has a high probability of rapidly progressing to frankly malignant MM with osteolytic lesions and/or an increasing tumor mass. Further progression of MM is associated with increasingly severe secondary features (lytic bone lesions, anemia, immunodeficiency, renal impairment), and in some patients the occurrence of tumor in extramedullary locations. Extramedullary MM is a more aggressive tumor that often is called secondary or primary plasma cell leukemia (PCL), depending on whether or not preceding intramedullary myeloma has been recognized. Human MM cell lines (HMCL), which can be viewed as the ultimate stage of tumor progression, sometimes can be generated, but usually only from extramedullary tumors.
Ig Translocations Are Present in a Majority of Multiple Myeloma Tumors
Many B cell tumors have chromosomal translocations that involve the IgH locus (14q32) or one of the IgL loci (kappa, 2p12 and lambda, 22q11), and usually are mediated by errors in VDJ recombination or one of the other two B-cell-specific DNA modification mechanisms (above).13 The consequence of these translocations is dysregulation or increased expression of an oncogene that is positioned near one or more of the strong Ig enhancers. Translocations involving an IgH switch region uniquely dissociate the intronic and 3′ IgH enhancers, so that an oncogene might be juxtaposed to an IgH enhancer on each of the derivative chromosomes, as first demonstrated for FGFR3 on der(14) and MMSET on der(4) in MM.14 The prevalence of IgH translocations varies somewhat with the disease stage: nearly 50% in MGUS or SMM, 55%–73% in intramedullary MM, 85% in primary PCL, and > 90% in HMCL.7,15–17
Primary Translocations Involving Five Recurrent Partners
Five recurrent chromosomal partners (oncogenes) are involved in IgH translocations in MGUS and MM: 4p16 (MMSET and usually FGFR3), 6p21 (CYCLIN D3), 11q13 (CYCLIN D1), 16q23 (c-MAF), and 20q11 (MAFB). Together, the combined prevalence of these five IgH translocation partners is about 40% in MM, with approximately 15% 4p16, 3% 6p21, 15% 11q13, 5% 16q23, and 2% 20q11. The mostly simple reciprocal translocations involving the five recurrent translocation partners appear to be primary translocations that usually are mediated by errors in IgH switch recombination, but perhaps sometimes by errors in somatic hypermutation, during the maturation of B cells in germinal centers.14 Although poorly understood, it has been shown that the prevalence of t(11;14) is markedly increased in primary amyloidosis and in the rare MM tumors that express IgM or are nonsecretory, whereas the prevalence of t(4;14) is significantly increased in MM tumors that express IgA.17–19 The decreased prevalence of IgH translocations involving 4p16 and 16q23 in MGUS suggests that these translocations can cause de novo MM and/or are associated with rapid progression from MGUS to MM.
MYC Translocations as a Paradigm for Secondary (Ig) Translocations in MM
About 3% of MM tumors have secondary IgH translocations that target c-MYC at 8q24.20 Secondary translocations that dysregulate a MYC gene (c- >> N- > L-) by juxtaposing it to an Ig locus (IgH ~ Igl >> Igk) or to one of many other poorly characterized chromosomal loci are late progression events.7,21 The MYC translocations are absent or rare in MGUS but occur in 15% of MM tumors, 45% of advanced tumors, and 90% of HMCL. These translocations are not mediated by the B-cell-specific DNA modification mechanisms, which are inactive in normal or tumor PC. In contrast to the primary translocations described above, these secondary events often include unbalanced and complex translocations and insertions that can involve three chromosomes, sometimes with associated amplification, duplication, inversion, or deletion.
Other Ig Translocations in MM
Other IgH translocation partners have been identified in approximately 20% of MGUS and MM tumors.15–17 These other partners, which are poorly characterized, appear to be mostly non-recurrent or rare. These translocations seem to share the structural complexity and lack of IgH switch region involvement observed for MYC translocations, suggesting that they usually represent secondary translocations, which can occur at any time during tumor progression, including MGUS. Translocations involving an Igλ locus occur in about 10% of MGUS tumors, and approximately 20% of advanced MM tumors or HMCL.7,15 Trans-locations involving an Igκ locus are rare, occurring in only a few percent of MM tumors. Nearly half of IgL translocations in advanced MM tumors or HMCL target a MYC gene. Significantly, although all HMCL analyzed have either an IgH or IgL translocation, approximately 30% of MM tumors and 45% of MGUS tumors do not have either an IgH or IgL translocation. Surprisingly, however, two independent Ig translocations have been found in 5% of MGUS tumors, 25% of advanced MM tumors, and 58% of HMCL, consistent with an accumulation of secondary Ig translocations during tumor progression.15
Loss of Chromosome 13 Sequences: A Frequent Karyotypic Abnormality
Abnormal karyotypes are rarely obtained for MGUS tumors, but are identified in roughly one third of MM tumors, with the prevalence of abnormal karyotypes increasing as MM tumors become more proliferative and/or less stromal cell dependent.22 However, interphase FISH studies show that all MGUS and MM tumors have numeric and/or structural chromosome abnormalities. Loss of one copy of chromosome 13 or, less often, selective loss of 13q/13q14 sequences was one of the first chromosomal abnormalities that was found to be associated with a poor prognosis. It is of much graver prognostic significance when identified by conventional cytogenetics then by interphase FISH alone, perhaps explained in part by the proliferative potential implied by obtaining an abnormal metaphase in vitro. Loss of chromosome 13 sequences occurs in more than 50% of MM tumors and somewhat less often in MGUS. It affects all of the cells in most MM tumors and many MGUS, but some studies indicate that loss of chromosome 13 sequences is more likely to affect only a subset of MGUS tumor cells.23 Strikingly, loss of chromosome 13 sequences is found in approximately 90% of MGUS or MM tumors that have a t(4;14) or t(14;16) translocation, but has a much lower prevalence in MM tumors that have a t(11;14) translocation, an IgH translocation with an unknown partner, or no IgH translocation.16,17,24
Hyperdiploid and Non-Hyperdiploid Tumors
A number of years ago, it was reported that hypodiploid tumors have a poorer prognosis than hyperdiploid tumors.25 Nearly half of MM tumors are hyperdiploid (HRD) (48–75 chromosomes), and often have multiple trisomies involving eight odd chromosomes (3,5,7,9,11,15,19,21). Non-hyperdiploid (NHRD) tumors (< 48 or > 75 chromosomes) can be hypodiploid, pseudodiploid or subtetraploid, with clonal subtetraploid and hypodiploid cells often present in the same tumor. The five recurrent IgH translocations, which occur only infrequently in HRD tumors, are found in approximately 70% of NHRD tumors.26,27 Secondary translocations, which appear to include all MYC translocations, most—if not all—IgL translocations, most IgH translocations not involving the five recurrent partners, and some IgH translocations involving the five recurrent partners, seem to occur with a similar prevalence in HRD and NHRD tumors. However, the prevalence of all structural chromosomal abnormalities is nearly twice as high in NHRD compared to HRD tumors. Loss of chromosome 13 sequences occurs in 72% of NHRD tumors but only 37% of HRD tumors, which is explained partially by the increased prevalence of t(4;14) and t(14;16) in NHRD tumors. Initially, it was suggested that hypodiploidy is the critical prognostic factor and that loss of chromosome 13 sequences has no additional prognostic significance.26 However, this is an issue that remains to be clarified since studies from another group indicate that hypodiploidy and loss of chromosome 13 sequences can independently confer a poor prognosis in MM.28
Dysregulation of CYCLIN D1, 2, or 3: A Unifying, Early Oncogenic Event in MM and MGUS
Most tumor cells in MGUS and MM appear more similar to normal, non-proliferating PC than to normal, but highly proliferating PB, for which 30% or more of the cells can be in S phase. Surprisingly, however, despite a very low proliferation index, the level of cyclin D1, cyclin D2, or cyclin D3 mRNA in virtually all MM and MGUS tumors is relatively high, comparable to the level of cyclin D2 mRNA expressed in normal proliferating PB, and distinctly higher than in normal BM PC (Figure 2 ).29 About 25% of MGUS or MM tumors have an IgH translocation that directly dysregulates CYCLIN D1 (11q13), CYCLIN D3 (6p21), or a MAF gene (c-MAF, 16q23 or MAFB, 20q11) encoding a transcription factor that targets CYCLIN D2. Most MM tumors with a t(4; 14) translocation express high levels of cyclin D2, but usually at a level that is somewhat lower than tumors with a translocation that targets MAF or MAFB; there is no information about the mechanism causing increased cyclin D2 expression in the t(4;14) tumors. Although normal BM PC express little or no detectable cyclin D1, nearly 40% of MGUS and MM tumors do not have a t(11;14), but are hyperdiploid, have multiple trisomies of the eight odd chromosomes, and bi-allelically express increased levels of cyclin D1. Most other tumors, about half of which are hyperdiploid and have multiple trisomies of the eight odd chromosomes, show increased expression of cyclin D2 compared to normal BM PC.
A Model for the Molecular Pathogenesis of MGUS and Multiple Myeloma
The current model has been updated from an earlier version.7 It has been proposed that there are two pathways of pathogenesis: an NHRD pathway that usually includes one of the five recurrent IgH translocations as an early event, and a HRD pathway that is associated with multiple trisomies of eight odd chromosomes but is mediated by a yet to be determined mechanism.25,27 As summarized above, dysregulation of a CYCLIN D gene—sometimes as a consequence of a primary IgH translocation but otherwise by presently unknown mechanisms—appears to be a unifying and early event. The low proliferative capacity of MGUS or MM tumors that express a dysregulated CYCLIN D gene is consistent with the fact that a high level of transgenic CYCLIN D1 does not perturb normal B cell development and proliferation or lead to tumors, unless there is a cooperating MYC or activated RAS transgene. The dysregulation of a CYCLIN D gene may render the cells more susceptible to proliferative stimuli, resulting in selective expansion as a result of interaction with BM stromal cells that produce interleukin (IL)-6, insulin-like growth factor (IGF)-1, and other cytokines. Loss of chromosome 13/13q sequences also seems to be an early event shared by MGUS and MM tumors. Unfortunately, we do not yet fully understand the relative timing of primary IgH translocations, aneuploidy (including multiple trisomies and loss of chromosome 13 sequences), and CYCLIN D dysregulation. Secondary chromosome translocations and other karyotypic abnormalities can occur at all stages of tumorigenesis. Epigenetic changes, including methylation of promoter regions that can inactivate p16INK4a, p15INK4b, and other genes, also can occur at all stages of tumorigenesis.30 Mutually exclusive activating mutations of K- or N-RAS (or FGFR3 when there is a t(4;14) translocation) are rare in MGUS, but the prevalence of RAS mutations is 30%–40% in early MM and slightly higher in advanced MM; FGFR3 mutations appear to occur more frequently in advanced MM. Secondary MYC translocations are late progression events that may occur as a tumor becomes less dependent on BM stromal cells and/or more proliferative. Despite alteration of the RB pathway by dysregulation of a CYCLIN D gene in virtually all MGUS and MM tumors, inactivation of an additional component of this pathway (p18INK4c or RB) can be a late progression event that is associated with enhanced proliferation. Mutations and/or mono-allelic deletion of p53 also appear to be late progression events. The timing of other events, such as PTEN mutations, is unknown.
A TC (Translocation/Cyclin D Expression) Classification Based on Early Pathogenic Events
Based in large part on the hypotheses presented above, a supervised analysis of gene expression profiles provides the basis for a molecular classification of MM. In addition to determining the expression level of cyclin D1, 2, and 3, gene expression profiling can effectively identify MM tumors that overexpress the oncogenes dysregulated by the five recurrent IgH translocations: 11q13 (CYCLIN D1); 6p21 (CYCLIN D3); 4p16 (MMSET & usually FGFR3); 16q23 (c-MAF); and 20q11 (MAFB).29 These groups (Table 1 ) can be distinguished based on the Ig translocation present, and cyclin D expression: 11q13 (16%) and 6p21 tumors (3%) express high levels of either cyclin D1 or cyclin D3 as a result of an Ig translocation; D1 tumors (34%) ectopically express low to moderate levels of cyclin D1 despite the absence of a t(11;14) translocation; D1+D2 (6%) in addition express cyclin D2. D2 tumors (17%), which are a mixture of hyperdiploid and non-hyperdiploid tumors that do not fall into one of the other groups, express increased cyclin D2 compared to normal PC. Tumors in group None (2%) do not have increased expression of a D-type cyclin compared to normal bone marrow PC. 4p16 tumors (15%) express high levels of cyclin D2, and also MMSET (and FGFR3 in approximately 70%) as a result of a t(4;14) translocation; maf tumors (7%) express the highest levels of cyclin D2, and also high levels of either c-maf or mafB, consistent with the possibility that both maf transcription factors up-regulate the expression of cyclin D2. Supervised hierarchical cluster analysis of gene expression profiles demonstrates that the TC classification identifies homogeneous groups of tumors with distinctive patterns of gene expression, and by corollary, phenotype. Although not unequivocally established, we think that the basis for assignment of tumors to the TC groups is focused primarily on very early if not initiating oncogenic events that are shared by MGUS and MM tumors, although the D1+D2 group might represent an exception.
Implications of the TC Classification of MGUS and MM
In addition to having shared gene expression profiles, we have identified important biologic and clinical correlates associated with the TC groups (Table 1 ). For example, the TC D1 group of tumors is absent or under-represented in PCL and HMCL, suggesting that these tumors have a particularly strong dependence on a continued interaction with bone marrow stromal cells. In addition, we have found that lytic bone disease correlates with the TC classification, with high prevalence (~90%) in TC 6p21, TC 11q13, TC D1 and TC D1+D2, and lower prevalence (~55%) in TC 4p16 and TC maf. It has also become clear that specific IgH translocations have a profound prognostic significance.31,32 Patients with tumors that have a t(4;14) translocation (TC 4p16) have a substantially shortened survival either with standard or high-dose therapy (median OS 26 months and 33 months respectively), and patients with a t(14;16) (TC maf) have a similarly poor if not worse prognosis (median OS 16 months with conventional therapy). By contrast, patients with tumors that have a t(11;14) translocation (TC 11q13) appear to have a better survival following both conventional chemotherapy and high-dose therapy.33 Similarly we suspect that the TC D1 group, representing most of the hyperdiploid patients, shares the good prognosis associated with hyperdiploidy. There are too few patients to draw conclusions about TC 6p21 but given the overlapping gene expression profile with TC 11q13 and obvious mechanistic similarities, it makes sense to group them together. Similarly it makes sense to group the t(14;16) (c-maf) with t(14;20) (mafB) into the TC maf group. Although we do not have mature data at this time we suspect that D1+D2, which have a higher proliferative index, and are overrepresented in relapsed patients, have a poor prognosis. The TC NONE group is very small, but as it represents patients with macrofocal disease it would appear to have a good prognosis. These results suggest that the TC classification, which appears to be based on the earliest events in pathogenesis, may be a clinically useful way to classify patients into groups that have distinct subtypes of MM (and MGUS) tumors.16,18,34,35 One might argue that some or all of the TC groups represent different disease that may require different therapeutic approaches. Although the TC groups, which are based on what appear to be early—perhaps initiating—pathogenic events, provide a foundation for clinically relevant insights, it is likely that a definitive molecular classification will require modification as additional initiating and progression events are identified.
ConcludingThoughts Regarding Prognosis and Treatment of MM
Two general kinds of parameters have been used to assess the prognosis and response to treatment of MM: a combination of tumor mass plus host response, and intrinsic properties of the tumor cell.2,16,25,36–39 Clinical indicators of host response, often related to the mass of tumor, include anemia, thrombocytopenia, lytic bone disease, immunodeficiency, and compromised renal function. Tumor mass is measured most directly by the fraction of tumor cells in bone marrow aspirates or biopsies, or less directly by the level of serum M-Ig or urine M-IgL, whereas additional measures of host response can include serum levels of polyclonal Ig, hemoglobin, β2-microglobulin, lactic dehydrogenase, and C-reactive protein. Regarding intrinsic properties of the tumor cell, it has been shown that an unfavorable outcome is associated with each of the following: increased plasma cell labeling index (PCLI) or an increased fraction of Ki67 positive (cycling) cells, an abnormal karyotype, hypodiploidy, loss of chromosome 13/13q sequences, monosomy of 17/p53 deletion, loss of 1p sequences, and gain of 1q sequences. It also has been reported that mutations of K-RAS (but not N-RAS) represent an adverse prognostic factor. With the recent development and application of new genomic technologies (gene expression profiling, array comparative genomic hybridization, single nucleotide polymorphism analysis), there will be an increasing trend to develop and evaluate therapies based on patient specific genotypic and phenotypic properties of both the tumor cells and normal cells. In the short run, these recent advances will help to more precisely classify MM tumors that respond most favorably to one or more of an ever-widening variety of therapeutic regimens. In the not too far distant future, however, we are hopeful that our rapidly increasing understanding of the molecular and cellular biology of MM, will lead to therapies that directly target different kinds of MM tumor cells (such as FGFR3 inhibitors for tumors with t[4;14]),40 or their requisite interaction with the host microenvironment. We think that this is a time to be optimistic that the rapid pace of MM research has brought us close to having the potential of identifying therapies that will either cure MM or convert MM to a chronic disease in a significant fraction of patients with this presently incurable malignancy.41,42
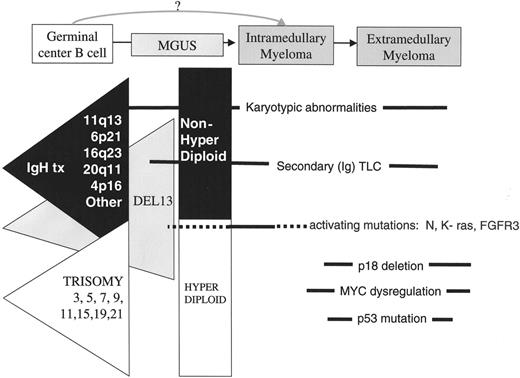
Disease stages and timing of oncogenic events. The earliest oncogenic changes are present in monoclonal gammopathy of undetermined significance (MGUS) and involve two minimally overlapping pathways, primary IgH translocations (black triangle) and multiple trisomies (white triangle), each of which can include a del 13 pathway (grey triangle). Other karyotypic abnormalities, including secondary (Ig) TLC, and epigenetic changes can occur at all stages. Activating mutations of K- or N-RAS appear to mark, if not cause, the MGUS to multiple myeloma (MM) transition in some cases, but sometimes occur during subsequent progression of MM. Late oncogenic events that occur at a time when tumors are becoming more aggressive include MYC dysregulation by secondary (Ig) TLC, bi-allelic deletion of p18, inactivation of Rb, and loss or mutation of p53.
Disease stages and timing of oncogenic events. The earliest oncogenic changes are present in monoclonal gammopathy of undetermined significance (MGUS) and involve two minimally overlapping pathways, primary IgH translocations (black triangle) and multiple trisomies (white triangle), each of which can include a del 13 pathway (grey triangle). Other karyotypic abnormalities, including secondary (Ig) TLC, and epigenetic changes can occur at all stages. Activating mutations of K- or N-RAS appear to mark, if not cause, the MGUS to multiple myeloma (MM) transition in some cases, but sometimes occur during subsequent progression of MM. Late oncogenic events that occur at a time when tumors are becoming more aggressive include MYC dysregulation by secondary (Ig) TLC, bi-allelic deletion of p18, inactivation of Rb, and loss or mutation of p53.
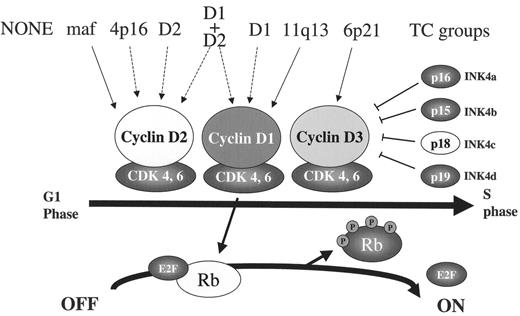
Alteration of RB pathway by both early and late pathogenic events. An early pathogenic event in tumors from seven of the translocation and cyclin D (TC) groups is dysregulation of one of the three CYCLIN D genes, either as a consequence of an Ig TLC (solid arrow), or by an unknown mechanism (dashed arrow). Increased expression of one of the Cyclin D proteins facilitates activation of CDK4 (or CDK6), which then phosphorylates and inactivates Rb so that E2F can facilitate G1>S cell cycle progression. This reaction is regulated by CDK inhibitors (INK4a-d), so that increased proliferation of some multiple myeloma (MM) tumors occurs only after a late oncogenic event that inactivates Rb or p18INK4c.
Alteration of RB pathway by both early and late pathogenic events. An early pathogenic event in tumors from seven of the translocation and cyclin D (TC) groups is dysregulation of one of the three CYCLIN D genes, either as a consequence of an Ig TLC (solid arrow), or by an unknown mechanism (dashed arrow). Increased expression of one of the Cyclin D proteins facilitates activation of CDK4 (or CDK6), which then phosphorylates and inactivates Rb so that E2F can facilitate G1>S cell cycle progression. This reaction is regulated by CDK inhibitors (INK4a-d), so that increased proliferation of some multiple myeloma (MM) tumors occurs only after a late oncogenic event that inactivates Rb or p18INK4c.
WMK: Genetics Branch, National Cancer Institute, Bethesda, MD; PLB: Mayo Clinic, Comprehensive Cancer Center and Division of Hematology-Oncology, Scottsdale, AZ
Supported in part by NIH grant CA100707 (PLB)